Making upset plots from GHRU data
Anthony Underwood
plotting_upset_diagrams.RmdIntroduction
UpSet diagrams visualizes complex intersections where the columns represent different combinations of factors across different sets of groups (rows). They are very helpful when looking at the different combinations of AMR determinants that may be responsible for resistance.
Getting and formatting the data
First get the relevant data sets
library(ghruR)
library(kableExtra)
epi_data <- ghruR::get_data_for_country(
country_value = "India",
type_value = "Epidemiological Metadata",
user_email = "anthony.underwood@cgps.group")## [1] "anthony.underwood@cgps.group"
## [1] "anthony.underwood@cgps.group"
## [1] "anthony.underwood@cgps.group"
epi_data <- ghruR::clean_data(epi_data)
ast_data <- ghruR::get_data_for_country(
country_value = "India",
type_value = "Antimicrobial Susceptibility Testing",
user_email = "anthony.underwood@cgps.group")## [1] "anthony.underwood@cgps.group"
acquired_amr_data <- ghruR::get_data_for_country(
country_value = "India",
type_value = "AMR Klebsiella pneumoniae",
AMR_type = "acquired",
user_email = "anthony.underwood@cgps.group")## [1] "anthony.underwood@cgps.group"
point_amr_data <- ghruR::get_data_for_country(
country_value = "India",
type_value = "AMR Klebsiella pneumoniae",
AMR_type = "point",
user_email = "anthony.underwood@cgps.group")## [1] "anthony.underwood@cgps.group"
mlst_data <- ghruR::get_data_for_country(
country_value = "India",
type_value = "MLST",
user_email = "anthony.underwood@cgps.group")## [1] "anthony.underwood@cgps.group"Parsing the acquired AMR data
Convert the acquired AMR data to long format, annotate it and get the different data sets
# convert to long format
annotated_amr_data <- ghruR::annotate_amr_data(acquired_amr_data)
# filter data for those that have acquired the gene and have good pct_id match and ctg_cov
acquired_genes_present <- ghruR::filter_long_data(annotated_amr_data)
# filter by subclass
carbapenamases <- acquired_genes_present %>%
dplyr::filter(subclass == "CARBAPENEM") %>%
dplyr::select(`Sample id`, gene) %>%
dplyr::rename(determinant = gene)
head(carbapenamases) %>% kable() %>% kable_styling() %>% scroll_box(width = "100%")| Sample id | determinant |
|---|---|
| G18250048 | blaNDM-1 |
| G18250048 | blaOXA-181 |
| G18250048 | blaOXA-181 |
| G18250088 | blaNDM-1 |
| G18250088 | blaOXA-232 |
| G18250088 | blaOXA-232 |
esbls <- acquired_genes_present %>%
dplyr::filter(subclass == "CEPHALOSPORIN") %>%
dplyr::select(`Sample id`, gene) %>%
dplyr::rename(determinant = gene) %>%
dplyr::filter(
str_starts(determinant, "blaCTX-M") |
str_starts(determinant, "blaSHV") |
str_starts(determinant, "blaTEM") |
str_starts(determinant, "blaOXA")
)
head(esbls) %>% kable() %>% kable_styling() %>% scroll_box(width = "100%")| Sample id | determinant |
|---|---|
| G18250048 | blaCTX-M-15 |
| G18250088 | blaCTX-M-15 |
| G18250088 | blaOXA-1 |
| G18250103 | blaCTX-M-15 |
| G18250103 | blaOXA-1 |
| G18250114 | blaCTX-M-15 |
Parsing the point AMR data
Now handling point data, first get the mutations in assembled genes
annotated_point_amr_data <- ghruR::get_annotated_point_amr_data(point_amr_data, 'klebsiella', annotation_type = 'mutations')
# get mutations specific for carbapenems
carbapenam_specific_mutations <- annotated_point_amr_data %>%
dplyr::filter(resistance == "carbapenem") %>%
select(-resistance)
head(carbapenam_specific_mutations) %>% kable() %>% kable_styling() %>% scroll_box(width = "100%")| Sample id | determinant |
|---|---|
| G18250048 | ompK36_A217S |
| G18250048 | ompK37_E244D |
| G18250048 | ompK37_I128M |
| G18250048 | ompK37_I70M |
| G18250048 | ompK37_N230G |
| G18250088 | ompK36_A217S |
Now get the genes which were incomplete (partial or fragmented) and therefore possibly non-functional
annotated_point_amr_data <- ghruR::get_annotated_point_amr_data(point_amr_data, 'klebsiella', annotation_type = 'incomplete')
# get mutations specific for carbapenems
carbapenam_specific_incomplete_genes <- annotated_point_amr_data %>%
dplyr::filter(resistance == "carbapenem") %>%
select(-resistance)
head(carbapenam_specific_incomplete_genes) %>% kable() %>% kable_styling() %>% scroll_box(width = "100%")| Sample id | determinant |
|---|---|
| G18251889 | ompK36-absent |
| G18252053 | ompK36-absent |
| G18252054 | ompK36-partial |
| G18253166 | ompK36-partial |
| G18253170 | ompK36-absent |
| G18253171 | ompK36-partial |
For simplicity in the plots convert both of these to be either gene-mutation or gene-incomplete
carbapenam_specific_mutations %<>% dplyr::mutate(determinant = stringr::str_replace(determinant, "_.+$", "-mutation")) %>%
dplyr::distinct(`Sample id`, determinant)
head(carbapenam_specific_mutations) %>% kable() %>% kable_styling() %>% scroll_box(width = "100%")| Sample id | determinant |
|---|---|
| G18250048 | ompK36-mutation |
| G18250048 | ompK37-mutation |
| G18250088 | ompK36-mutation |
| G18250088 | ompK37-mutation |
| G18250103 | ompK36-mutation |
| G18250103 | ompK37-mutation |
carbapenam_specific_incomplete_genes %<>% dplyr::mutate(determinant = stringr::str_replace(determinant, "-.+$", "-incomplete")) %>%
dplyr::distinct(`Sample id`, determinant)
head(carbapenam_specific_incomplete_genes) %>% kable() %>% kable_styling() %>% scroll_box(width = "100%")| Sample id | determinant |
|---|---|
| G18251889 | ompK36-incomplete |
| G18252053 | ompK36-incomplete |
| G18252054 | ompK36-incomplete |
| G18253166 | ompK36-incomplete |
| G18253170 | ompK36-incomplete |
| G18253171 | ompK36-incomplete |
Combining all the dataframes
Produce a final dataframe from the acquired and point dataframes with determinants putatively involved in carbapenem resistance
This involves binding the rows and combining multiple determinants for the same sample into one by joining them into a list (for the ggupset function) using the group_by and summarise functions from the the dplyr package
#combine all determinants
all_potential_CR_determinants <-
rbind(carbapenamases, esbls, carbapenam_specific_mutations, carbapenam_specific_incomplete_genes)
# merge multiple instances
all_potential_CR_determinants %<>% dplyr::group_by(`Sample id`) %>%
dplyr::summarise(determinant = list(determinant))
head(all_potential_CR_determinants) %>% kable() %>% kable_styling() %>% scroll_box(width = "100%")| Sample id | determinant |
|---|---|
| G18250048 | blaNDM-1 , blaOXA-181 , blaOXA-181 , blaCTX-M-15 , ompK36-mutation, ompK37-mutation |
| G18250088 | blaNDM-1 , blaOXA-232 , blaOXA-232 , blaCTX-M-15 , blaOXA-1 , ompK36-mutation, ompK37-mutation |
| G18250103 | blaNDM-1 , blaOXA-232 , blaOXA-232 , blaCTX-M-15 , blaOXA-1 , ompK36-mutation, ompK37-mutation |
| G18250104 | ompK36-mutation, ompK37-mutation |
| G18250114 | blaNDM-1 , blaCTX-M-15 , blaOXA-1 , ompK36-mutation, ompK37-mutation |
| G18250122 | blaNDM-1 , blaCTX-M-15 , blaOXA-1 , ompK36-mutation, ompK37-mutation |
Filtering out samples without relevant loci
- filter out those without a carbapenamase
- Filter out those that have no beta lactamase and those that have a potential ESBL without a porin mutation
all_potential_CR_determinants %<>%
dplyr::filter(str_detect(determinant, "bla"))
# filter out those with only ESBL but no OmpK mutations
all_potential_CR_determinants %<>%
dplyr::filter(!
(
(
str_detect(determinant, "blaCTX-M") |
str_detect(determinant, "blaSHV") |
str_detect(determinant, "blaTEM") |
str_detect(determinant, "blaOXA")
) &
!str_detect(determinant, "ompK")
)
)
head(all_potential_CR_determinants) %>% kable() %>% kable_styling() %>% scroll_box(width = "100%")| Sample id | determinant |
|---|---|
| G18250048 | blaNDM-1 , blaOXA-181 , blaOXA-181 , blaCTX-M-15 , ompK36-mutation, ompK37-mutation |
| G18250088 | blaNDM-1 , blaOXA-232 , blaOXA-232 , blaCTX-M-15 , blaOXA-1 , ompK36-mutation, ompK37-mutation |
| G18250103 | blaNDM-1 , blaOXA-232 , blaOXA-232 , blaCTX-M-15 , blaOXA-1 , ompK36-mutation, ompK37-mutation |
| G18250114 | blaNDM-1 , blaCTX-M-15 , blaOXA-1 , ompK36-mutation, ompK37-mutation |
| G18250122 | blaNDM-1 , blaCTX-M-15 , blaOXA-1 , ompK36-mutation, ompK37-mutation |
| G18251889 | blaOXA-232 , blaOXA-232 , blaCTX-M-15 , ompK37-mutation , ompK36-incomplete |
Plotting the data
Use the ggupset library to plot the data
library(ggupset)
# Find unique determinants to become the set labels
unique_determinants <- rbind(carbapenamases, esbls, carbapenam_specific_mutations, carbapenam_specific_incomplete_genes) %>%
dplyr::distinct(determinant) %>%
dplyr::arrange(determinant) %>%
dplyr::pull(determinant)
# make the upset plot
upset_plot <- all_potential_CR_determinants %>%
ggplot(aes(x=determinant)) +
geom_bar() +
scale_x_upset(order_by = "freq", sets = unique_determinants)
upset_plot
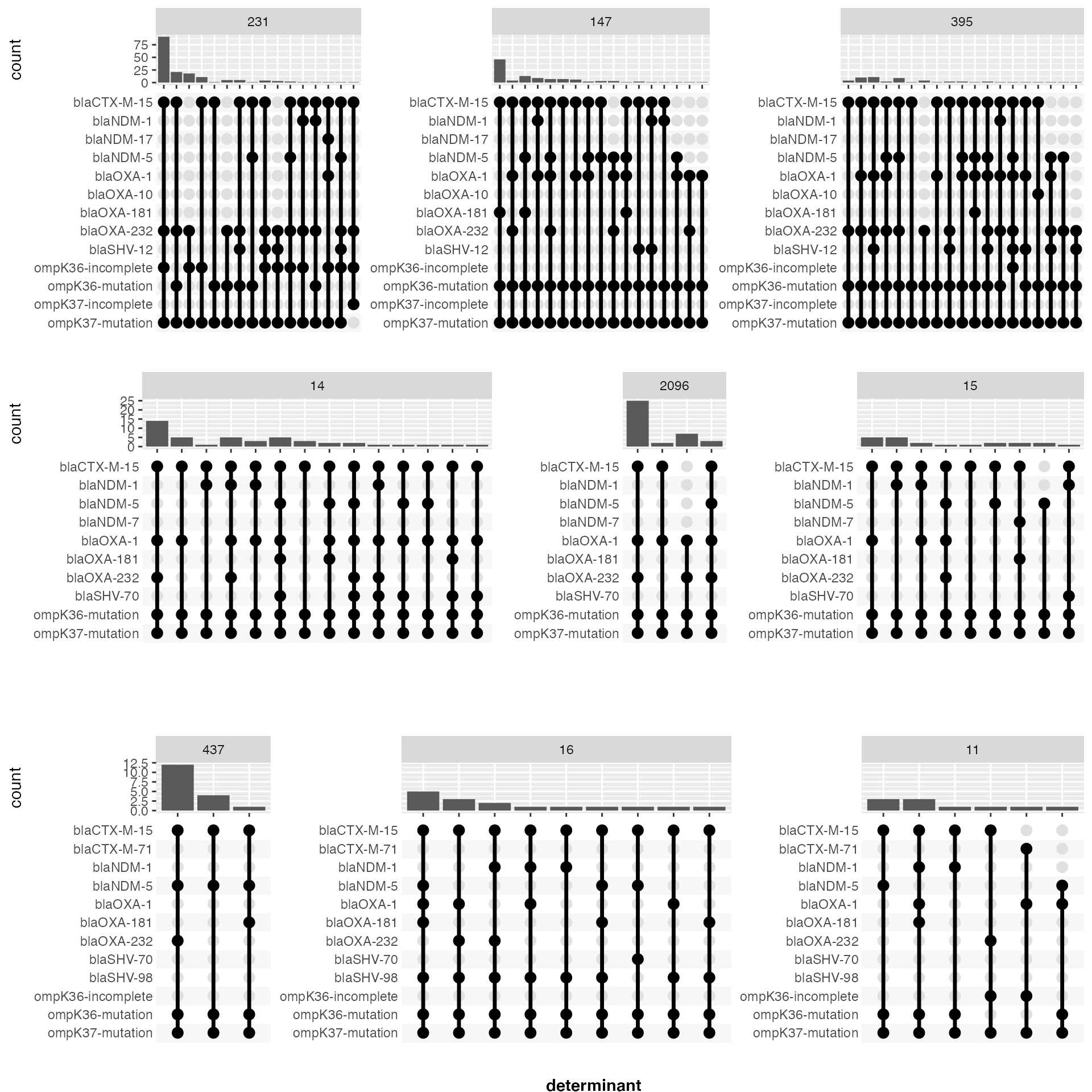
Breaking it down by ST
library(cowplot)
library(forcats)
all_potential_CR_determinants_and_ST <- all_potential_CR_determinants %>%
dplyr::left_join(mlst_data %>% dplyr::select(`Sample id`, ST), by = "Sample id")
num <- 9
most_frequent_STs <- all_potential_CR_determinants_and_ST %>%
dplyr::select(ST) %>% dplyr::count(ST) %>%
dplyr::arrange(desc(n)) %>% dplyr::slice_max(n, n = num) %>%
dplyr::pull(ST)
plots = list()
plot_index <- 0
for (start in seq(1,9, by =3)){
plot_index <- plot_index + 1
end <- start + 2
selected_STs <- most_frequent_STs[start:end]
data_for_STs <- all_potential_CR_determinants_and_ST %>% dplyr::filter(ST %in% selected_STs)
determinants_for_STs <- data_for_STs %>%
unnest(cols = c(determinant)) %>%
dplyr::distinct(determinant) %>%
dplyr::arrange(determinant) %>%
dplyr::pull(determinant)
upset_plot_for_STs <- data_for_STs %>%
mutate(ST = fct_relevel(ST, selected_STs)) %>%
ggplot(aes(x = determinant)) +
geom_bar() +
theme(plot.title = element_text(size = 20, face = "bold")) +
facet_grid(cols = vars(ST), scales = "free_x", space = "free", drop = FALSE) +
ggeasy::easy_labs(x = "") +
scale_x_upset(order_by = "freq", sets = determinants_for_STs) +
theme(panel.spacing.x=unit(6, "lines"))
plots[[plot_index]] <- upset_plot_for_STs
}
plot_grid <- cowplot::plot_grid(plotlist = plots, align = "h", ncol = 1) +
cowplot::draw_figure_label(label = "determinant", position = "bottom", font = "bold")
plot_grid