Making Microreact-compatible data from GHRU metadata the manual way
Anthony Underwood
making_microreact_compatible_data_manually.RmdIntroduction
For more fine controlled over the data here are a few approaches
Combining Epi and acquired AMR data
Load libraries
library(ghruR)
library(kableExtra)Fetch the amr data
amr_data <- ghruR::get_data_for_country(
country_value = "India",
type_value = "AMR Klebsiella pneumoniae",
AMR_type = "acquired",
user_email = "anthony.underwood@cgps.group")
[1] "anthony.underwood@cgps.group"
[1] "anthony.underwood@cgps.group"
[1] "anthony.underwood@cgps.group"Use the ghruR function to replace the ariba cluster names with gene names or a common prefix based on the gene name and pivot data to long format for filtering
annotated_amr_data <- annotate_amr_data(amr_data)
cleaned_amr_data <- replace_ariba_cluster_names(annotated_amr_data)Filter the data based on the assembly metrics. Default is * assembled starts with “yes” * pct_id >= 90 * ctg_cov >= 5 Once filtered, teh data is then transformed back to wide format and sort by ene name/prefix
filtered_long_amr_data <- filter_long_data(
cleaned_amr_data,
assembled_starts_with = "yes",
pct_id_cutoff = 90,
ctg_cov_cutoff = 5
) %>%
select(`Sample id`, amr_gene_column, gene) %>%
distinct()
# transform back to wide format
renamed_amr_data <- filtered_long_amr_data %>%
pivot_wider(id_cols = `Sample id`, names_from = amr_gene_column, values_from = gene)
# sort the columns
colnames_without_id <- sort(setdiff(colnames(renamed_amr_data), c('Sample id')))
renamed_amr_data %<>% dplyr::select(`Sample id`, all_of(colnames_without_id))
head(renamed_amr_data) %>% kable() %>% kable_styling() %>% scroll_box(width = "100%")| Sample id | aac(3)-IId;aac(3)-IIe | aac(6’)-Ib4;aac(6’)-Ib;aac(6’)-Ib-cr5 | aadA | aadA13 | aadA16 | aadA2 | aadA5 | ant(2’’)-Ia | aph(3’’)-Ib | aph(3’)-Ia | aph(3’)-VI;aph(3’)-VIb | aph(6)-Id | armA | arr-2;arr-3 | blaCMY-6 | blaCTX-M-8 | blaCTX-M-9 | blaDHA-1;blaDHA-7 | blaLAP-2 | blaNDM-1 | blaOXA-1;blaOXA-796 | blaOXA-10 | blaOXA-181 | blaOXA-4 | blaOXA-9 | blaSFO-1 | blaSHV | blaSHV-2 | blaTEM-1 | ble | catA1 | catA2 | catB | catB3 | cmlA5;cmlA4 | dfrA1 | dfrA12 | dfrA14 | dfrA15 | dfrA17 | dfrA27 | dfrA5;dfrA30 | ere(A) | erm(B) | floR2 | fosA10 | mph(A) | mph(E) | msr(E) | oqxA1 | oqxB1 | qacEdelta1;qacE | qepA4 | qnrA9;qnrA1 | qnrB1;qnrB7;qnrB9 | qnrB4 | qnrB95;qnrB6 | qnrS1;qnrS8;qnrS9 | rmtB1;rmtB4 | rmtC | rmtF1;rmtF2 | sat2;estX/sat2 | sul1 | sul2 | sul3 | tet(A) | tet(B) | tet(D) | tet(G) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G18250048 | aac(3)-IId | aac(6’)-Ib4 | NA | NA | NA | NA | NA | NA | aph(3’’)-Ib | NA | NA | aph(6)-Id | NA | arr-2 | blaCMY-6 | blaCTX-M-15 | NA | NA | NA | blaNDM-1 | NA | NA | blaOXA-181 | blaOXA-181 | NA | NA | NA | blaSHV-182 | blaTEM-1 | ble | catA1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | fosA6 | NA | NA | NA | oqxA6 | oqxB20 | qacEdelta1 | NA | NA | qnrB1 | NA | NA | qnrS1 | NA | rmtC | NA | NA | sul1 | sul2 | NA | NA | NA | NA | NA |
| G18250088 | aac(3)-IIe | aac(6’)-Ib4 | aadA1 | NA | NA | NA | NA | NA | aph(3’’)-Ib | NA | aph(3’)-VI | aph(6)-Id | NA | arr-2 | NA | blaCTX-M-15 | NA | NA | NA | blaNDM-1 | blaOXA-1 | NA | blaOXA-232 | blaOXA-232 | blaOXA-9 | NA | blaSHV-89 | NA | blaTEM-219 | ble | NA | NA | catB | NA | NA | NA | NA | dfrA14 | NA | NA | NA | NA | NA | NA | NA | fosA6 | NA | NA | NA | NA | NA | NA | NA | NA | qnrB1 | NA | NA | qnrS1 | NA | NA | rmtF1 | NA | NA | sul2 | NA | tet(A) | NA | NA | NA |
| G18250103 | aac(3)-IIe | NA | aadA1 | NA | NA | NA | NA | NA | aph(3’’)-Ib | NA | aph(3’)-VI | aph(6)-Id | NA | arr-2 | NA | blaCTX-M-15 | NA | NA | NA | blaNDM-1 | blaOXA-1 | NA | blaOXA-232 | blaOXA-232 | blaOXA-9 | NA | blaSHV-89 | NA | blaTEM-90 | ble | NA | NA | catB | NA | NA | NA | NA | dfrA14 | NA | NA | NA | NA | NA | NA | NA | fosA6 | NA | NA | NA | NA | NA | NA | NA | NA | qnrB1 | NA | NA | qnrS1 | NA | NA | rmtF1 | NA | NA | sul2 | NA | tet(A) | NA | NA | NA |
| G18250104 | NA | aac(6’)-Ib | aadA1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | blaOXA-9 | NA | NA | blaSHV-67 | blaTEM-90 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | fosA | NA | NA | NA | oqxA7 | oqxB19 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| G18250114 | NA | NA | aadA1 | NA | NA | NA | NA | NA | NA | NA | aph(3’)-VI | NA | NA | arr-3 | NA | blaCTX-M-15 | NA | NA | NA | blaNDM-1 | blaOXA-1 | NA | NA | NA | blaOXA-9 | NA | NA | blaSHV-67 | blaTEM-90 | ble | NA | NA | NA | catB3 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | fosA | NA | NA | NA | oqxA7 | oqxB19 | qacEdelta1 | NA | NA | NA | NA | NA | qnrS1 | NA | NA | NA | NA | sul1 | NA | NA | NA | NA | NA | NA |
| G18250122 | NA | NA | aadA1 | NA | NA | NA | NA | NA | NA | NA | aph(3’)-VI | NA | NA | arr-3 | NA | blaCTX-M-15 | NA | NA | NA | blaNDM-1 | blaOXA-1 | NA | NA | NA | blaOXA-9 | NA | NA | blaSHV-67 | blaTEM-90 | ble | NA | NA | NA | catB3 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | fosA | NA | NA | NA | oqxA7 | oqxB19 | qacEdelta1 | NA | NA | NA | NA | NA | qnrS1 | NA | NA | NA | NA | sul1 | NA | NA | NA | NA | NA | NA |
Add colour to the data
# NA means not available in R and in this case the NAs mean the gene was not detected so let's replace `NA` with `-`
renamed_amr_data %<>% mutate(across(everything(), ~replace_na(.x, '-')))
coloured_renamed_amr_data <- ghruR::colour_dataframe(
renamed_amr_data,
id_column = 'Sample id',
absent_colour = 'grey',
present_colour = 'red',
absence_character = '-'
)
head(coloured_renamed_amr_data) %>% kable() %>% kable_styling() %>% scroll_box(width = "100%")| Sample id | aac(3)-IId;aac(3)-IIe | aac(3)-IId;aac(3)-IIe__colour | aac(6’)-Ib4;aac(6’)-Ib;aac(6’)-Ib-cr5 | aac(6’)-Ib4;aac(6’)-Ib;aac(6’)-Ib-cr5__colour | aadA | aadA__colour | aadA13 | aadA13__colour | aadA16 | aadA16__colour | aadA2 | aadA2__colour | aadA5 | aadA5__colour | ant(2’’)-Ia | ant(2’’)-Ia__colour | aph(3’’)-Ib | aph(3’’)-Ib__colour | aph(3’)-Ia | aph(3’)-Ia__colour | aph(3’)-VI;aph(3’)-VIb | aph(3’)-VI;aph(3’)-VIb__colour | aph(6)-Id | aph(6)-Id__colour | armA | armA__colour | arr-2;arr-3 | arr-2;arr-3__colour | blaCMY-6 | blaCMY-6__colour | blaCTX-M-8 | blaCTX-M-8__colour | blaCTX-M-9 | blaCTX-M-9__colour | blaDHA-1;blaDHA-7 | blaDHA-1;blaDHA-7__colour | blaLAP-2 | blaLAP-2__colour | blaNDM-1 | blaNDM-1__colour | blaOXA-1;blaOXA-796 | blaOXA-1;blaOXA-796__colour | blaOXA-10 | blaOXA-10__colour | blaOXA-181 | blaOXA-181__colour | blaOXA-4 | blaOXA-4__colour | blaOXA-9 | blaOXA-9__colour | blaSFO-1 | blaSFO-1__colour | blaSHV | blaSHV__colour | blaSHV-2 | blaSHV-2__colour | blaTEM-1 | blaTEM-1__colour | ble | ble__colour | catA1 | catA1__colour | catA2 | catA2__colour | catB | catB__colour | catB3 | catB3__colour | cmlA5;cmlA4 | cmlA5;cmlA4__colour | dfrA1 | dfrA1__colour | dfrA12 | dfrA12__colour | dfrA14 | dfrA14__colour | dfrA15 | dfrA15__colour | dfrA17 | dfrA17__colour | dfrA27 | dfrA27__colour | dfrA5;dfrA30 | dfrA5;dfrA30__colour | ere(A) | ere(A)__colour | erm(B) | erm(B)__colour | floR2 | floR2__colour | fosA10 | fosA10__colour | mph(A) | mph(A)__colour | mph(E) | mph(E)__colour | msr(E) | msr(E)__colour | oqxA1 | oqxA1__colour | oqxB1 | oqxB1__colour | qacEdelta1;qacE | qacEdelta1;qacE__colour | qepA4 | qepA4__colour | qnrA9;qnrA1 | qnrA9;qnrA1__colour | qnrB1;qnrB7;qnrB9 | qnrB1;qnrB7;qnrB9__colour | qnrB4 | qnrB4__colour | qnrB95;qnrB6 | qnrB95;qnrB6__colour | qnrS1;qnrS8;qnrS9 | qnrS1;qnrS8;qnrS9__colour | rmtB1;rmtB4 | rmtB1;rmtB4__colour | rmtC | rmtC__colour | rmtF1;rmtF2 | rmtF1;rmtF2__colour | sat2;estX/sat2 | sat2;estX/sat2__colour | sul1 | sul1__colour | sul2 | sul2__colour | sul3 | sul3__colour | tet(A) | tet(A)__colour | tet(B) | tet(B)__colour | tet(D) | tet(D)__colour | tet(G) | tet(G)__colour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G18250048 | aac(3)-IId | red | aac(6’)-Ib4 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | aph(3’’)-Ib | red |
|
grey |
|
grey | aph(6)-Id | red |
|
grey | arr-2 | red | blaCMY-6 | red | blaCTX-M-15 | red |
|
grey |
|
grey |
|
grey | blaNDM-1 | red |
|
grey |
|
grey | blaOXA-181 | red | blaOXA-181 | red |
|
grey |
|
grey |
|
grey | blaSHV-182 | red | blaTEM-1 | red | ble | red | catA1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | fosA6 | red |
|
grey |
|
grey |
|
grey | oqxA6 | red | oqxB20 | red | qacEdelta1 | red |
|
grey |
|
grey | qnrB1 | red |
|
grey |
|
grey | qnrS1 | red |
|
grey | rmtC | red |
|
grey |
|
grey | sul1 | red | sul2 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
| G18250088 | aac(3)-IIe | red | aac(6’)-Ib4 | red | aadA1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | aph(3’’)-Ib | red |
|
grey | aph(3’)-VI | red | aph(6)-Id | red |
|
grey | arr-2 | red |
|
grey | blaCTX-M-15 | red |
|
grey |
|
grey |
|
grey | blaNDM-1 | red | blaOXA-1 | red |
|
grey | blaOXA-232 | red | blaOXA-232 | red | blaOXA-9 | red |
|
grey | blaSHV-89 | red |
|
grey | blaTEM-219 | red | ble | red |
|
grey |
|
grey | catB | red |
|
grey |
|
grey |
|
grey |
|
grey | dfrA14 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | fosA6 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | qnrB1 | red |
|
grey |
|
grey | qnrS1 | red |
|
grey |
|
grey | rmtF1 | red |
|
grey |
|
grey | sul2 | red |
|
grey | tet(A) | red |
|
grey |
|
grey |
|
grey |
| G18250103 | aac(3)-IIe | red |
|
grey | aadA1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | aph(3’’)-Ib | red |
|
grey | aph(3’)-VI | red | aph(6)-Id | red |
|
grey | arr-2 | red |
|
grey | blaCTX-M-15 | red |
|
grey |
|
grey |
|
grey | blaNDM-1 | red | blaOXA-1 | red |
|
grey | blaOXA-232 | red | blaOXA-232 | red | blaOXA-9 | red |
|
grey | blaSHV-89 | red |
|
grey | blaTEM-90 | red | ble | red |
|
grey |
|
grey | catB | red |
|
grey |
|
grey |
|
grey |
|
grey | dfrA14 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | fosA6 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | qnrB1 | red |
|
grey |
|
grey | qnrS1 | red |
|
grey |
|
grey | rmtF1 | red |
|
grey |
|
grey | sul2 | red |
|
grey | tet(A) | red |
|
grey |
|
grey |
|
grey |
| G18250104 |
|
grey | aac(6’)-Ib | red | aadA1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | blaOXA-9 | red |
|
grey |
|
grey | blaSHV-67 | red | blaTEM-90 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | fosA | red |
|
grey |
|
grey |
|
grey | oqxA7 | red | oqxB19 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
| G18250114 |
|
grey |
|
grey | aadA1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | aph(3’)-VI | red |
|
grey |
|
grey | arr-3 | red |
|
grey | blaCTX-M-15 | red |
|
grey |
|
grey |
|
grey | blaNDM-1 | red | blaOXA-1 | red |
|
grey |
|
grey |
|
grey | blaOXA-9 | red |
|
grey |
|
grey | blaSHV-67 | red | blaTEM-90 | red | ble | red |
|
grey |
|
grey |
|
grey | catB3 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | fosA | red |
|
grey |
|
grey |
|
grey | oqxA7 | red | oqxB19 | red | qacEdelta1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | qnrS1 | red |
|
grey |
|
grey |
|
grey |
|
grey | sul1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
| G18250122 |
|
grey |
|
grey | aadA1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | aph(3’)-VI | red |
|
grey |
|
grey | arr-3 | red |
|
grey | blaCTX-M-15 | red |
|
grey |
|
grey |
|
grey | blaNDM-1 | red | blaOXA-1 | red |
|
grey |
|
grey |
|
grey | blaOXA-9 | red |
|
grey |
|
grey | blaSHV-67 | red | blaTEM-90 | red | ble | red |
|
grey |
|
grey |
|
grey | catB3 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | fosA | red |
|
grey |
|
grey |
|
grey | oqxA7 | red | oqxB19 | red | qacEdelta1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | qnrS1 | red |
|
grey |
|
grey |
|
grey |
|
grey | sul1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
library(ape)
tree <- read.tree("input_data/india_kpn.tre")
leaf_labels <- tree$tip.label
epi_data <- ghruR::get_data_for_country(
country_value = "India",
type_value = "Epidemiological Metadata",
AMR_type = "acquired",
user_email = "anthony.underwood@cgps.group")
[1] "anthony.underwood@cgps.group"
# function to clean data - this can be messy
epi_data <- clean_data(epi_data)
# include only the epi metadata that corresponds to the tree leaves
epi_data %<>% filter(`Sample id` %in% leaf_labels) %>%
rename(id = `Sample id`)
# add row for ref
ref_dataframe <- data.frame('id' = 'NC_016845.1')
epi_data_with_ref <- plyr::rbind.fill(epi_data, ref_dataframe)Combine with amr data
microreact_metadata <- epi_data_with_ref %>%
left_join(coloured_renamed_amr_data, by = c('id' = 'Sample id'))
# remove nas and add empty string instead. It looks cleaner
microreact_metadata %<>% mutate(across(everything(), ~replace_na(.x, '')))
# This dataframe is ready to be written to a CSV for microreact
write_csv(microreact_metadata, 'output_data/single_country_microreact_manual.csv')
head(microreact_metadata) %>% kable() %>% kable_styling() %>% scroll_box(width = "100%")| id | Alternative sample id | GHRU UUID | Species | Country | Country alpha-2 Code | Region/Province/Department | ISO 3166-2 subdivision code | Location | Sentinel Site Code | Latitude | Longitude | Collected by | Collection Contact | Year | Month | Day | Outbreak isolate | Clinical significance | Host | Specimen type | Invasive | Patient type | Ward (Inpatients Only) | Room (Inpatients Only) | Address (Outpatients only) | Address Latitude (Outpatients only) | Address Longitude (Outpatients only) | Origin | HAI Type | Patient Date of Birth | Patient age | Patient gender | Clinical Diagnosis | Collection date | aac(3)-IId;aac(3)-IIe | aac(3)-IId;aac(3)-IIe__colour | aac(6’)-Ib4;aac(6’)-Ib;aac(6’)-Ib-cr5 | aac(6’)-Ib4;aac(6’)-Ib;aac(6’)-Ib-cr5__colour | aadA | aadA__colour | aadA13 | aadA13__colour | aadA16 | aadA16__colour | aadA2 | aadA2__colour | aadA5 | aadA5__colour | ant(2’’)-Ia | ant(2’’)-Ia__colour | aph(3’’)-Ib | aph(3’’)-Ib__colour | aph(3’)-Ia | aph(3’)-Ia__colour | aph(3’)-VI;aph(3’)-VIb | aph(3’)-VI;aph(3’)-VIb__colour | aph(6)-Id | aph(6)-Id__colour | armA | armA__colour | arr-2;arr-3 | arr-2;arr-3__colour | blaCMY-6 | blaCMY-6__colour | blaCTX-M-8 | blaCTX-M-8__colour | blaCTX-M-9 | blaCTX-M-9__colour | blaDHA-1;blaDHA-7 | blaDHA-1;blaDHA-7__colour | blaLAP-2 | blaLAP-2__colour | blaNDM-1 | blaNDM-1__colour | blaOXA-1;blaOXA-796 | blaOXA-1;blaOXA-796__colour | blaOXA-10 | blaOXA-10__colour | blaOXA-181 | blaOXA-181__colour | blaOXA-4 | blaOXA-4__colour | blaOXA-9 | blaOXA-9__colour | blaSFO-1 | blaSFO-1__colour | blaSHV | blaSHV__colour | blaSHV-2 | blaSHV-2__colour | blaTEM-1 | blaTEM-1__colour | ble | ble__colour | catA1 | catA1__colour | catA2 | catA2__colour | catB | catB__colour | catB3 | catB3__colour | cmlA5;cmlA4 | cmlA5;cmlA4__colour | dfrA1 | dfrA1__colour | dfrA12 | dfrA12__colour | dfrA14 | dfrA14__colour | dfrA15 | dfrA15__colour | dfrA17 | dfrA17__colour | dfrA27 | dfrA27__colour | dfrA5;dfrA30 | dfrA5;dfrA30__colour | ere(A) | ere(A)__colour | erm(B) | erm(B)__colour | floR2 | floR2__colour | fosA10 | fosA10__colour | mph(A) | mph(A)__colour | mph(E) | mph(E)__colour | msr(E) | msr(E)__colour | oqxA1 | oqxA1__colour | oqxB1 | oqxB1__colour | qacEdelta1;qacE | qacEdelta1;qacE__colour | qepA4 | qepA4__colour | qnrA9;qnrA1 | qnrA9;qnrA1__colour | qnrB1;qnrB7;qnrB9 | qnrB1;qnrB7;qnrB9__colour | qnrB4 | qnrB4__colour | qnrB95;qnrB6 | qnrB95;qnrB6__colour | qnrS1;qnrS8;qnrS9 | qnrS1;qnrS8;qnrS9__colour | rmtB1;rmtB4 | rmtB1;rmtB4__colour | rmtC | rmtC__colour | rmtF1;rmtF2 | rmtF1;rmtF2__colour | sat2;estX/sat2 | sat2;estX/sat2__colour | sul1 | sul1__colour | sul2 | sul2__colour | sul3 | sul3__colour | tet(A) | tet(A)__colour | tet(B) | tet(B)__colour | tet(D) | tet(D)__colour | tet(G) | tet(G)__colour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G18250048 | NIHR/JOD/KPN-67 | KPN | India | IN | Rajasthan | IN-RJ | JODHPUR | AIIMSJ | 26.2389 | 73.0243 | AIIMS | Dr. Anuradha Sharma | 2017 | 3 | 14 | Human | ur | 1 | Male | aac(3)-IId | red | aac(6’)-Ib4 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | aph(3’’)-Ib | red |
|
grey |
|
grey | aph(6)-Id | red |
|
grey | arr-2 | red | blaCMY-6 | red | blaCTX-M-15 | red |
|
grey |
|
grey |
|
grey | blaNDM-1 | red |
|
grey |
|
grey | blaOXA-181 | red | blaOXA-181 | red |
|
grey |
|
grey |
|
grey | blaSHV-182 | red | blaTEM-1 | red | ble | red | catA1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | fosA6 | red |
|
grey |
|
grey |
|
grey | oqxA6 | red | oqxB20 | red | qacEdelta1 | red |
|
grey |
|
grey | qnrB1 | red |
|
grey |
|
grey | qnrS1 | red |
|
grey | rmtC | red |
|
grey |
|
grey | sul1 | red | sul2 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | |||||||||||||||
| G18250088 | NIHR/JOD/KPN-70 | KPN | India | IN | Rajasthan | IN-RJ | JODHPUR | AIIMSJ | 26.2389 | 73.0243 | AIIMS | Dr. Anuradha Sharma | 2017 | 3 | 16 | Human | ur | 48 | Male | aac(3)-IIe | red | aac(6’)-Ib4 | red | aadA1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | aph(3’’)-Ib | red |
|
grey | aph(3’)-VI | red | aph(6)-Id | red |
|
grey | arr-2 | red |
|
grey | blaCTX-M-15 | red |
|
grey |
|
grey |
|
grey | blaNDM-1 | red | blaOXA-1 | red |
|
grey | blaOXA-232 | red | blaOXA-232 | red | blaOXA-9 | red |
|
grey | blaSHV-89 | red |
|
grey | blaTEM-219 | red | ble | red |
|
grey |
|
grey | catB | red |
|
grey |
|
grey |
|
grey |
|
grey | dfrA14 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | fosA6 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | qnrB1 | red |
|
grey |
|
grey | qnrS1 | red |
|
grey |
|
grey | rmtF1 | red |
|
grey |
|
grey | sul2 | red |
|
grey | tet(A) | red |
|
grey |
|
grey |
|
grey | |||||||||||||||
| G18250103 | NIHR/JOD/KPN-72 | KPN | India | IN | Rajasthan | IN-RJ | JODHPUR | AIIMSJ | 26.2389 | 73.0243 | AIIMS | Dr. Anuradha Sharma | 2017 | 3 | 16 | Human | bl | 5 | Male | aac(3)-IIe | red |
|
grey | aadA1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | aph(3’’)-Ib | red |
|
grey | aph(3’)-VI | red | aph(6)-Id | red |
|
grey | arr-2 | red |
|
grey | blaCTX-M-15 | red |
|
grey |
|
grey |
|
grey | blaNDM-1 | red | blaOXA-1 | red |
|
grey | blaOXA-232 | red | blaOXA-232 | red | blaOXA-9 | red |
|
grey | blaSHV-89 | red |
|
grey | blaTEM-90 | red | ble | red |
|
grey |
|
grey | catB | red |
|
grey |
|
grey |
|
grey |
|
grey | dfrA14 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | fosA6 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | qnrB1 | red |
|
grey |
|
grey | qnrS1 | red |
|
grey |
|
grey | rmtF1 | red |
|
grey |
|
grey | sul2 | red |
|
grey | tet(A) | red |
|
grey |
|
grey |
|
grey | |||||||||||||||
| G18250104 | NIHR/JOD/KPN-76 | KPN | India | IN | Rajasthan | IN-RJ | JODHPUR | AIIMSJ | 26.2389 | 73.0243 | AIIMS | Dr. Anuradha Sharma | 2017 | 7 | 4 | Human | ur | 53 | Female |
|
grey | aac(6’)-Ib | red | aadA1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | blaOXA-9 | red |
|
grey |
|
grey | blaSHV-67 | red | blaTEM-90 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | fosA | red |
|
grey |
|
grey |
|
grey | oqxA7 | red | oqxB19 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | |||||||||||||||
| G18250114 | NIHR/JOD/KPN-77 | KPN | India | IN | Rajasthan | IN-RJ | JODHPUR | AIIMSJ | 26.2389 | 73.0243 | AIIMS | Dr. Anuradha Sharma | 2017 | 8 | 4 | Human | ur | 48 | Male |
|
grey |
|
grey | aadA1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | aph(3’)-VI | red |
|
grey |
|
grey | arr-3 | red |
|
grey | blaCTX-M-15 | red |
|
grey |
|
grey |
|
grey | blaNDM-1 | red | blaOXA-1 | red |
|
grey |
|
grey |
|
grey | blaOXA-9 | red |
|
grey |
|
grey | blaSHV-67 | red | blaTEM-90 | red | ble | red |
|
grey |
|
grey |
|
grey | catB3 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | fosA | red |
|
grey |
|
grey |
|
grey | oqxA7 | red | oqxB19 | red | qacEdelta1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | qnrS1 | red |
|
grey |
|
grey |
|
grey |
|
grey | sul1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | |||||||||||||||
| G18250122 | NIHR/JOD/KPN-79 | KPN | India | IN | Rajasthan | IN-RJ | JODHPUR | AIIMSJ | 26.2389 | 73.0243 | AIIMS | Dr. Anuradha Sharma | 2017 | 4 | 14 | Human | ur | 39 | Male |
|
grey |
|
grey | aadA1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | aph(3’)-VI | red |
|
grey |
|
grey | arr-3 | red |
|
grey | blaCTX-M-15 | red |
|
grey |
|
grey |
|
grey | blaNDM-1 | red | blaOXA-1 | red |
|
grey |
|
grey |
|
grey | blaOXA-9 | red |
|
grey |
|
grey | blaSHV-67 | red | blaTEM-90 | red | ble | red |
|
grey |
|
grey |
|
grey | catB3 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | fosA | red |
|
grey |
|
grey |
|
grey | oqxA7 | red | oqxB19 | red | qacEdelta1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | qnrS1 | red |
|
grey |
|
grey |
|
grey |
|
grey | sul1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
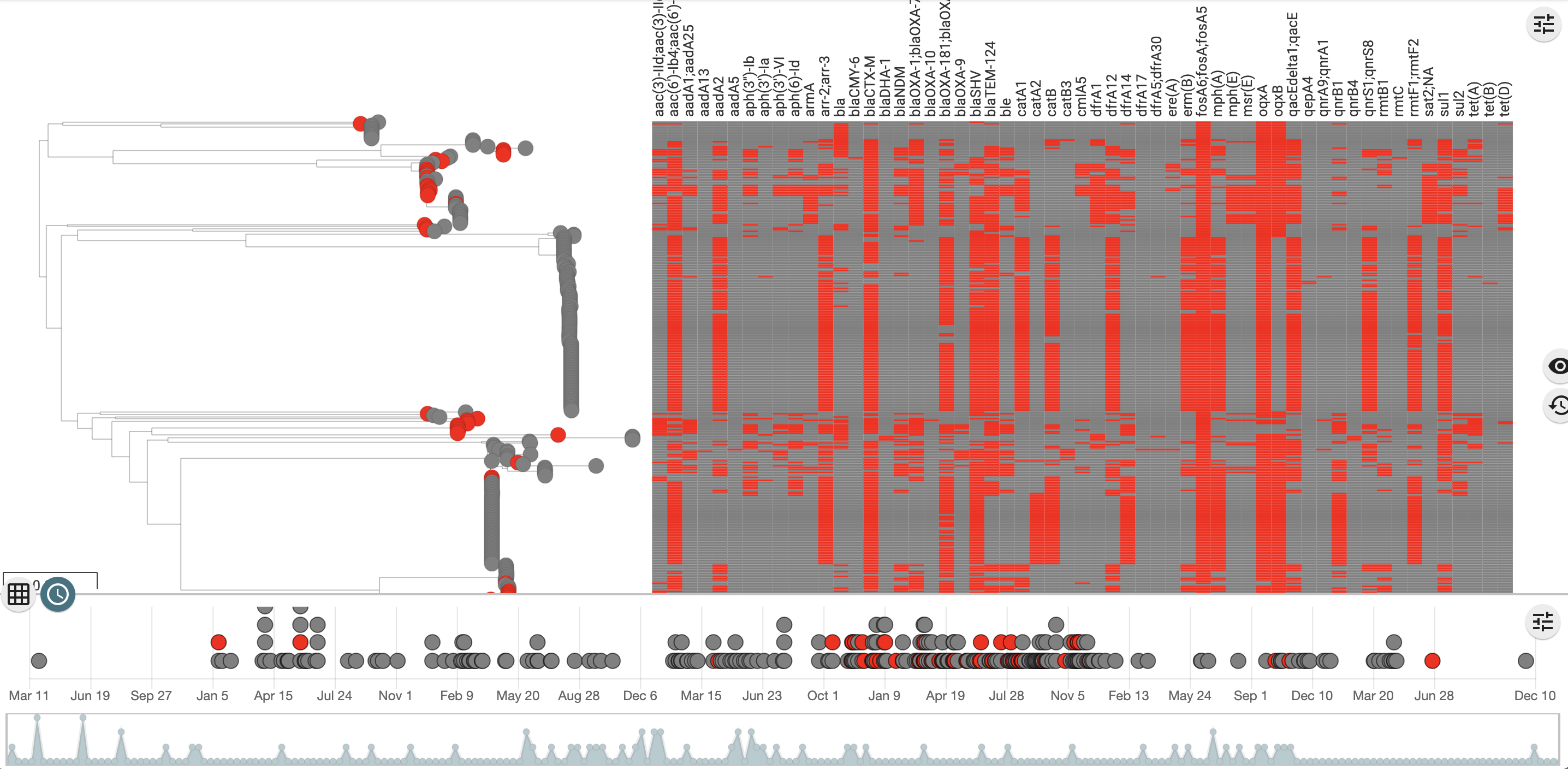
Producing summary columns instead
It may sometimes be preferrable to group the columns by drug class and just display if there is a determinant present that has been associated with the class. For the acquired genes the NCBI subclasses have many different variants and some should be rationalised. A suggestion would be to use an external table as a source for combining. Here is an example hosted in a Google Sheet 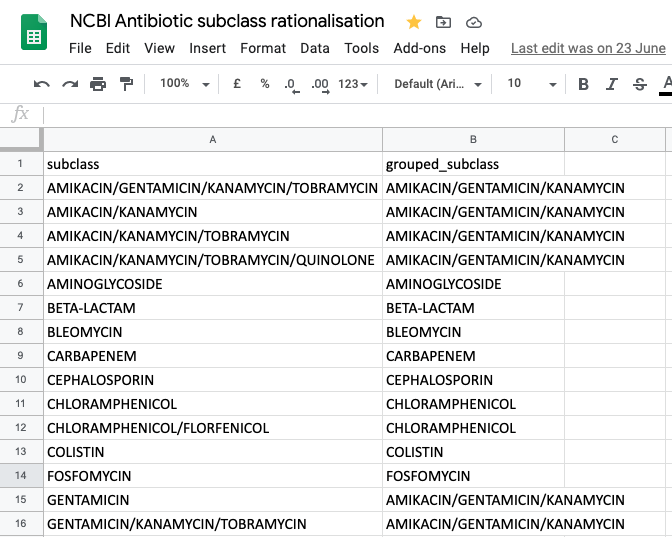 This data can be used to group the subclasses. We’ll start with the annotated_acquired amr_data
This data can be used to group the subclasses. We’ll start with the annotated_acquired amr_data
annotated_amr_data <- ghruR::annotate_amr_data(amr_data)
# fetch the subclass groups
subclass_groups <- googlesheets4::read_sheet("https://docs.google.com/spreadsheets/d/1X4-xD2-e78geJOFLOKri2QjeOzhrPLHzwsAN_f8mX1Q")
# use this to merge subclasses
annotated_amr_data %<>%
drop_na(refseq_nucleotide_accession) %>%
dplyr::left_join(subclass_groups %>% select(-class), by = "subclass") %>%
dplyr::select(-subclass) %>%
dplyr::rename(subclass = grouped_subclass)
head(annotated_amr_data) %>% kable() %>% kable_styling() %>% scroll_box(width = "100%")| Sample id | ariba_cluster | assembled | refseq_nucleotide_accession | pct_id | ctg_cov | gene_family | product_name | class | gene | subclass |
|---|---|---|---|---|---|---|---|---|---|---|
| G18250048 | A7J11_01233+ | partial | NG_047213.1 | 99.64 | 28.1 | aac(6’)-30/aac(6’)-Ib’ | bifunctional aminoglycoside N-acetyltransferase AAC(6’)-30/aminoglycoside N-acetyltransferase AAC(6’)-Ib’ | AMINOGLYCOSIDE | aac(6’)-30/aac(6’)-Ib’ | NA |
| G18250048 | aac_3__II- | yes | NG_047251.1 | 100.0 | 43.8 | aac(3)-IId | aminoglycoside N-acetyltransferase AAC(3)-IId | AMINOGLYCOSIDE | aac(3)-IId | AMIKACIN/GENTAMICIN/KANAMYCIN |
| G18250048 | aac_6___Ib+ | yes | NG_051844.1 | 99.82 | 27.7 | aac(6’)-Ib | aminoglycoside N-acetyltransferase AAC(6’)-Ib4 | AMINOGLYCOSIDE | aac(6’)-Ib4 | AMIKACIN/GENTAMICIN/KANAMYCIN |
| G18250048 | aph_3____Ib | yes | NG_056002.2 | 100.0 | 36.0 | aph(3’’)-Ib | aminoglycoside O-phosphotransferase APH(3’’)-Ib | AMINOGLYCOSIDE | aph(3’’)-Ib | STREPTOMYCIN |
| G18250048 | aph_6__Id | yes | NG_047464.1 | 100.0 | 23.3 | aph(6)-Id | aminoglycoside O-phosphotransferase APH(6)-Id | AMINOGLYCOSIDE | aph(6)-Id | STREPTOMYCIN |
| G18250048 | arr- | yes | NG_048580.1 | 100.0 | 35.2 | arr | NAD(+)–rifampin ADP-ribosyltransferase Arr-2 | RIFAMYCIN | arr-2 | RIFAMYCIN |
Sometimes the class doesn’t have a subclass. Therefore these columns should be combined using the coalesce funtion again.
annotated_amr_data %<>% dplyr::mutate(class = dplyr::coalesce(subclass, class)) %>%
select(`Sample id`,gene,class)
head(annotated_amr_data) %>% kable() %>% kable_styling() %>% scroll_box(width = "100%")| Sample id | gene | class |
|---|---|---|
| G18250048 | aac(6’)-30/aac(6’)-Ib’ | AMINOGLYCOSIDE |
| G18250048 | aac(3)-IId | AMIKACIN/GENTAMICIN/KANAMYCIN |
| G18250048 | aac(6’)-Ib4 | AMIKACIN/GENTAMICIN/KANAMYCIN |
| G18250048 | aph(3’’)-Ib | STREPTOMYCIN |
| G18250048 | aph(6)-Id | STREPTOMYCIN |
| G18250048 | arr-2 | RIFAMYCIN |
Many samples will have more than one determinant for some classes. These should be combined before returning the data frame to wide format of one row per sample.
In the final data we will want something for every sample even if it’s empty. This is the purpose of the final left join
# Use group_by and summarise functions to collapse multiple occurences of genes in the same class
annotated_amr_data %<>% dplyr::group_by(`Sample id`, class) %>%
dplyr::summarise(gene = paste0(gene, collapse = ','))
# pivot this data back to wide format
wide_annotated_amr_data <- annotated_amr_data %>%
drop_na(class) %>%
pivot_wider(names_from = c(class), values_from =c (gene))
# ensure all samples have data
wide_annotated_amr_data <- amr_data %>% dplyr::select(`Sample id`) %>%
dplyr::left_join(wide_annotated_amr_data, by = "Sample id")
head(wide_annotated_amr_data) %>% kable() %>% kable_styling() %>% scroll_box(width = "100%")| Sample id | AMIKACIN/GENTAMICIN/KANAMYCIN | AMINOGLYCOSIDE | BETA-LACTAM | BLEOMYCIN | CARBAPENEM | CEPHALOSPORIN | FOSFOMYCIN | MACROLIDE | PHENICOL | QUATERNARY AMMONIUM | QUINOLONE | RIFAMYCIN | STREPTOMYCIN | SULFONAMIDE | TETRACYCLINE | TRIMETHOPRIM | AMINOGLYCOSIDE/PHENICOL | AMINOGLYCOSIDE/QUINOLONE | STREPTOTHRICIN | ERYTHROMYCIN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G18250048 | aac(3)-IId,aac(6’)-Ib4 | aac(6’)-30/aac(6’)-Ib’,rmtC,aac(6’)-30/aac(6’)-Ib’ | blaSHV-182,blaOXA-731,blaTEM-1 | ble | blaNDM-1,blaOXA-181,blaOXA-181 | blaCMY-6,blaCTX-M-15 | fosA6 | ere(A) | catA1,oqxA6,oqxB20 | qacEdelta1 | qnrB1,qnrS1 | arr-2 | aph(3’’)-Ib,aph(6)-Id | sul1,sul2 | NA | NA | NA | NA | NA | NA |
| G18250088 | aac(3)-IIe,aac(6’)-Ib4,aph(3’)-VI | aac(6’)-30/aac(6’)-Ib’,rmtF1,aac(6’)-30/aac(6’)-Ib’ | blaSHV-89,blaOXA-9,blaTEM-219,blaSHV-89,blaOXA-9 | ble | blaNDM-1,blaOXA-232,blaOXA-232 | blaCTX-M-15,blaOXA-1 | fosA6,fosA7.4 | ere(A) | catB,catB3 | NA | qnrB1,qnrS1 | arr-2 | aadA13,aadA1,aph(3’’)-Ib,aph(6)-Id | sul2 | tet(A) | dfrA14 | NA | NA | NA | NA |
| G18250103 | aac(3)-IIe,aac(6’)-Ib,aph(3’)-VI | aac(6’)-30/aac(6’)-Ib’,rmtF1,aac(6’)-30/aac(6’)-Ib’ | blaSHV-89,blaOXA-9,blaTEM-90,blaSHV-89,blaOXA-9 | ble | blaNDM-1,blaOXA-232,blaOXA-232 | blaCTX-M-15,blaOXA-1 | fosA6,fosA7.4 | ere(A) | catB,catB3 | NA | qnrB1,qnrS1 | arr-2 | aadA13,aadA1,aph(3’’)-Ib,aph(6)-Id | sul2 | tet(A) | dfrA14 | NA | NA | NA | NA |
| G18250104 | aac(6’)-Ib | aac(6’)-30/aac(6’)-Ib’,aac(6’)-30/aac(6’)-Ib’ | blaSHV-67,blaOXA-9,blaTEM-90,blaOXA-9 | NA | NA | NA | fosA | NA | oqxA7,oqxB19 | NA | NA | NA | aadA13,aadA1 | NA | NA | NA | catB8/aac(6’)-Ib’,catB8/aac(6’)-Ib’ | NA | NA | NA |
| G18250114 | aac(6’)-Ib,aph(3’)-VI | aac(6’)-30/aac(6’)-Ib’,aac(6’)-30/aac(6’)-Ib’ | blaSHV-67,blaOXA-9,blaTEM-90,blaOXA-9 | ble | blaNDM-1 | blaCTX-M-15,blaOXA-1 | fosA | NA | catB3,oqxA7,oqxB19 | qacEdelta1 | qnrS1 | arr-3 | aadA13,aadA1 | sul1 | NA | NA | NA | NA | NA | NA |
| G18250122 | aac(6’)-Ib,aph(3’)-VI | aac(6’)-30/aac(6’)-Ib’,aac(6’)-30/aac(6’)-Ib’ | blaSHV-67,blaOXA-9,blaTEM-90,blaOXA-9 | ble | blaNDM-1 | blaCTX-M-15,blaOXA-1 | fosA | NA | catB3,oqxA7,oqxB19 | qacEdelta1 | qnrS1 | arr-3 | aadA13,aadA1 | sul1 | NA | NA | NA | NA | NA | NA |
Finally we’ll colour and join it with the epi data as above
# NA means not available in R and in this case the NAs mean the gene was not detected so let's replace `NA` with `-`
wide_annotated_amr_data %<>% mutate(across(everything(), ~replace_na(.x, '-')))
coloured_clustered_amr_data <- ghruR::colour_dataframe(
wide_annotated_amr_data,
id_column = 'Sample id',
absent_colour = 'grey',
present_colour = 'red',
absence_character = '-'
)
microreact_metadata <- epi_data_with_ref %>%
left_join(coloured_clustered_amr_data, by = c('id' = 'Sample id'))
# remove nas and add empty string instead. It looks cleaner
microreact_metadata %<>% mutate(across(everything(), ~replace_na(.x, '')))
# This dataframe is ready to be written to a CSV for microreact
write_csv(microreact_metadata, 'output_data/single_country_microreact_clustered_manual.csv')
head(microreact_metadata) %>% kable() %>% kable_styling() %>% scroll_box(width = "100%")| id | Alternative sample id | GHRU UUID | Species | Country | Country alpha-2 Code | Region/Province/Department | ISO 3166-2 subdivision code | Location | Sentinel Site Code | Latitude | Longitude | Collected by | Collection Contact | Year | Month | Day | Outbreak isolate | Clinical significance | Host | Specimen type | Invasive | Patient type | Ward (Inpatients Only) | Room (Inpatients Only) | Address (Outpatients only) | Address Latitude (Outpatients only) | Address Longitude (Outpatients only) | Origin | HAI Type | Patient Date of Birth | Patient age | Patient gender | Clinical Diagnosis | Collection date | AMIKACIN/GENTAMICIN/KANAMYCIN | AMIKACIN/GENTAMICIN/KANAMYCIN__colour | AMINOGLYCOSIDE | AMINOGLYCOSIDE__colour | BETA-LACTAM | BETA-LACTAM__colour | BLEOMYCIN | BLEOMYCIN__colour | CARBAPENEM | CARBAPENEM__colour | CEPHALOSPORIN | CEPHALOSPORIN__colour | FOSFOMYCIN | FOSFOMYCIN__colour | MACROLIDE | MACROLIDE__colour | PHENICOL | PHENICOL__colour | QUATERNARY AMMONIUM | QUATERNARY AMMONIUM__colour | QUINOLONE | QUINOLONE__colour | RIFAMYCIN | RIFAMYCIN__colour | STREPTOMYCIN | STREPTOMYCIN__colour | SULFONAMIDE | SULFONAMIDE__colour | TETRACYCLINE | TETRACYCLINE__colour | TRIMETHOPRIM | TRIMETHOPRIM__colour | AMINOGLYCOSIDE/PHENICOL | AMINOGLYCOSIDE/PHENICOL__colour | AMINOGLYCOSIDE/QUINOLONE | AMINOGLYCOSIDE/QUINOLONE__colour | STREPTOTHRICIN | STREPTOTHRICIN__colour | ERYTHROMYCIN | ERYTHROMYCIN__colour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G18250048 | NIHR/JOD/KPN-67 | KPN | India | IN | Rajasthan | IN-RJ | JODHPUR | AIIMSJ | 26.2389 | 73.0243 | AIIMS | Dr. Anuradha Sharma | 2017 | 3 | 14 | Human | ur | 1 | Male | aac(3)-IId,aac(6’)-Ib4 | red | aac(6’)-30/aac(6’)-Ib’,rmtC,aac(6’)-30/aac(6’)-Ib’ | red | blaSHV-182,blaOXA-731,blaTEM-1 | red | ble | red | blaNDM-1,blaOXA-181,blaOXA-181 | red | blaCMY-6,blaCTX-M-15 | red | fosA6 | red | ere(A) | red | catA1,oqxA6,oqxB20 | red | qacEdelta1 | red | qnrB1,qnrS1 | red | arr-2 | red | aph(3’’)-Ib,aph(6)-Id | red | sul1,sul2 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | |||||||||||||||
| G18250088 | NIHR/JOD/KPN-70 | KPN | India | IN | Rajasthan | IN-RJ | JODHPUR | AIIMSJ | 26.2389 | 73.0243 | AIIMS | Dr. Anuradha Sharma | 2017 | 3 | 16 | Human | ur | 48 | Male | aac(3)-IIe,aac(6’)-Ib4,aph(3’)-VI | red | aac(6’)-30/aac(6’)-Ib’,rmtF1,aac(6’)-30/aac(6’)-Ib’ | red | blaSHV-89,blaOXA-9,blaTEM-219,blaSHV-89,blaOXA-9 | red | ble | red | blaNDM-1,blaOXA-232,blaOXA-232 | red | blaCTX-M-15,blaOXA-1 | red | fosA6,fosA7.4 | red | ere(A) | red | catB,catB3 | red |
|
grey | qnrB1,qnrS1 | red | arr-2 | red | aadA13,aadA1,aph(3’’)-Ib,aph(6)-Id | red | sul2 | red | tet(A) | red | dfrA14 | red |
|
grey |
|
grey |
|
grey |
|
grey | |||||||||||||||
| G18250103 | NIHR/JOD/KPN-72 | KPN | India | IN | Rajasthan | IN-RJ | JODHPUR | AIIMSJ | 26.2389 | 73.0243 | AIIMS | Dr. Anuradha Sharma | 2017 | 3 | 16 | Human | bl | 5 | Male | aac(3)-IIe,aac(6’)-Ib,aph(3’)-VI | red | aac(6’)-30/aac(6’)-Ib’,rmtF1,aac(6’)-30/aac(6’)-Ib’ | red | blaSHV-89,blaOXA-9,blaTEM-90,blaSHV-89,blaOXA-9 | red | ble | red | blaNDM-1,blaOXA-232,blaOXA-232 | red | blaCTX-M-15,blaOXA-1 | red | fosA6,fosA7.4 | red | ere(A) | red | catB,catB3 | red |
|
grey | qnrB1,qnrS1 | red | arr-2 | red | aadA13,aadA1,aph(3’’)-Ib,aph(6)-Id | red | sul2 | red | tet(A) | red | dfrA14 | red |
|
grey |
|
grey |
|
grey |
|
grey | |||||||||||||||
| G18250104 | NIHR/JOD/KPN-76 | KPN | India | IN | Rajasthan | IN-RJ | JODHPUR | AIIMSJ | 26.2389 | 73.0243 | AIIMS | Dr. Anuradha Sharma | 2017 | 7 | 4 | Human | ur | 53 | Female | aac(6’)-Ib | red | aac(6’)-30/aac(6’)-Ib’,aac(6’)-30/aac(6’)-Ib’ | red | blaSHV-67,blaOXA-9,blaTEM-90,blaOXA-9 | red |
|
grey |
|
grey |
|
grey | fosA | red |
|
grey | oqxA7,oqxB19 | red |
|
grey |
|
grey |
|
grey | aadA13,aadA1 | red |
|
grey |
|
grey |
|
grey | catB8/aac(6’)-Ib’,catB8/aac(6’)-Ib’ | red |
|
grey |
|
grey |
|
grey | |||||||||||||||
| G18250114 | NIHR/JOD/KPN-77 | KPN | India | IN | Rajasthan | IN-RJ | JODHPUR | AIIMSJ | 26.2389 | 73.0243 | AIIMS | Dr. Anuradha Sharma | 2017 | 8 | 4 | Human | ur | 48 | Male | aac(6’)-Ib,aph(3’)-VI | red | aac(6’)-30/aac(6’)-Ib’,aac(6’)-30/aac(6’)-Ib’ | red | blaSHV-67,blaOXA-9,blaTEM-90,blaOXA-9 | red | ble | red | blaNDM-1 | red | blaCTX-M-15,blaOXA-1 | red | fosA | red |
|
grey | catB3,oqxA7,oqxB19 | red | qacEdelta1 | red | qnrS1 | red | arr-3 | red | aadA13,aadA1 | red | sul1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey | |||||||||||||||
| G18250122 | NIHR/JOD/KPN-79 | KPN | India | IN | Rajasthan | IN-RJ | JODHPUR | AIIMSJ | 26.2389 | 73.0243 | AIIMS | Dr. Anuradha Sharma | 2017 | 4 | 14 | Human | ur | 39 | Male | aac(6’)-Ib,aph(3’)-VI | red | aac(6’)-30/aac(6’)-Ib’,aac(6’)-30/aac(6’)-Ib’ | red | blaSHV-67,blaOXA-9,blaTEM-90,blaOXA-9 | red | ble | red | blaNDM-1 | red | blaCTX-M-15,blaOXA-1 | red | fosA | red |
|
grey | catB3,oqxA7,oqxB19 | red | qacEdelta1 | red | qnrS1 | red | arr-3 | red | aadA13,aadA1 | red | sul1 | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
See this Microreact project to see how this looks 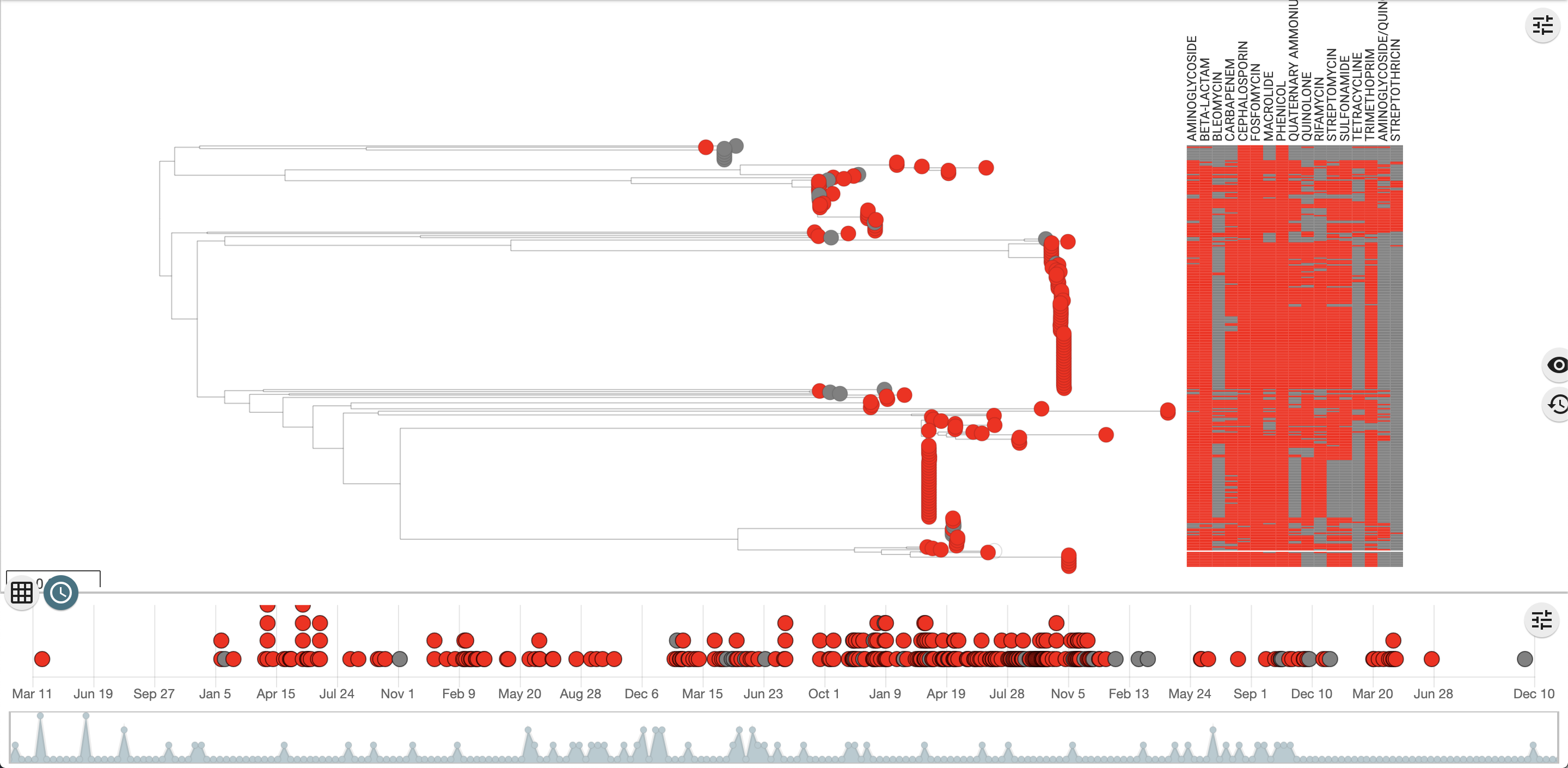
Making a Microreact metadata file with AMR genes from just one class
The subclass columns are very broad and sometimes it nay be helpful to split them out. Below is an example of how to make a microreact file with just carbapenemase genes
annotated_amr_data <- ghruR::annotate_amr_data(amr_data)
# fetch the subclass groups
subclass_groups <- googlesheets4::read_sheet("https://docs.google.com/spreadsheets/d/1X4-xD2-e78geJOFLOKri2QjeOzhrPLHzwsAN_f8mX1Q")
# use this to merge subclasses
annotated_amr_data %<>%
drop_na(refseq_nucleotide_accession) %>%
dplyr::left_join(subclass_groups %>% select(-class), by = "subclass") %>%
dplyr::select(-subclass) %>%
dplyr::rename(subclass = grouped_subclass)
carbapenem_related_genes <- annotated_amr_data %>%
filter(subclass == 'CARBAPENEM')
filtered_carbapenem_related_genes <- filter_long_data(
carbapenem_related_genes,
assembled_starts_with = "yes",
pct_id_cutoff = 90,
ctg_cov_cutoff = 5
) %>%
select(`Sample id`, gene, assembled) %>%
distinct()
# Make wide again
wide_carabapenem_related_genes <- filtered_carbapenem_related_genes %>% pivot_wider(id_cols = `Sample id`, names_from = 'gene', values_from = 'assembled')
# ensure all samples are present
wide_carabapenem_related_genes <- amr_data %>% dplyr::select(`Sample id`) %>%
dplyr::left_join(wide_carabapenem_related_genes, by = "Sample id") %>%
mutate(across(everything(), ~replace_na(.x, '-'))) %>%
arrange(`Sample id`) %>%
rename(id = `Sample id`)
# colour data
coloured_carbapenem_related_genes <- ghruR::colour_dataframe(
wide_carabapenem_related_genes,
id_column = 'id',
absent_colour = 'grey',
present_colour = 'red',
absence_character = '-'
)
# add row for ref
ref_dataframe <- data.frame('id' = 'NC_016845.1')
coloured_carbapenem_related_genes <- plyr::rbind.fill(coloured_carbapenem_related_genes, ref_dataframe)
head(coloured_carbapenem_related_genes) %>% kable() %>% kable_styling() %>% scroll_box(width = "100%")| id | blaNDM-1 | blaNDM-1__colour | blaOXA-181 | blaOXA-181__colour | blaOXA-232 | blaOXA-232__colour | blaNDM-5 | blaNDM-5__colour | blaNDM-17 | blaNDM-17__colour | blaNDM-7 | blaNDM-7__colour | blaOXA-48 | blaOXA-48__colour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G18250048 | yes | red | yes | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
| G18250088 | yes | red |
|
grey | yes | red |
|
grey |
|
grey |
|
grey |
|
grey |
| G18250103 | yes | red |
|
grey | yes | red |
|
grey |
|
grey |
|
grey |
|
grey |
| G18250104 |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
| G18250114 | yes | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
| G18250122 | yes | red |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
|
grey |
write_csv(coloured_carbapenem_related_genes, 'output_data/single_country_microreact_single_class_manual.csv')See this Microreact project to see how this looks 