Analysing GHRU Data
analysing_ghru_data.RmdGetting data from Google Spreadsheets
This requires some pre-requisites:
-
A Googlesheet (here we will call it a data type sheet list) with headers type and url that lists the urls of other googlesheets containing data. An example of the format is
type url Epidemiological Metadata https://docs.google.com/spreadsheets/d/123…. QC and Assembly Metadata https://docs.google.com/spreadsheets/d/456…. You can check the contents of a spreadsheet using the print_country_and_types command
library(ghruR)print_data_types( list_of_data_types_sheet_url = "https://docs.google.com/spreadsheets/d/1p0QfIZWQ55S5wZy9XQYn4L0kSUYMwLGPNYP5q9ef8kY", exclude_url = FALSE ) Warning: `data_frame()` was deprecated in tibble 1.1.0. Please use `tibble()` instead. This warning is displayed once every 8 hours. Call `lifecycle::last_lifecycle_warnings()` to see where this warning was generated. # A tibble: 6 × 2 type url <chr> <chr> 1 Antimicrobial Susceptibility Testing <NA> 2 Combined Data <NA> 3 Epidemiological Metadata https://docs.google.com/spreadsheets/d/1… 4 HiSeq X10 Sequencing Metadata <NA> 5 MiSeq Sequencing Metadata <NA> 6 QC and Assembly Metadata https://docs.google.com/spreadsheets/d/1… -
The linked spreadsheets contained in the data type sheet list contain the data that you can manipulate, merge, join and analyse. Therefore by specifying the data type the data frame containing that data can be fetched The available data types are:
- AMR Klebsiella pneumoniae
- Antimicrobial Susceptibility Testing
- Epidemiological Metadata
- HiSeq X10 Sequencing Metadata
- MiSeq Sequencing Metadata
- MLST
- QC and Assembly Metadata
Because each country has its own data we use a function that first retrieves the URL for a data type sheet list for the country specified and then gets the data for the data type specified To get the data from the
Epidemiological MetadataandQC and Assembly MetadatafromcountryXyou would use the following command for exampleepidemiological_metadata <- get_data_for_country('https://docs.google.com/spreadsheets/d/1D04XRov74Cw9LHgZTgkb4s8w4SfqbOOsjwD1k_UdQ-c', user_email = 'anthony.underwood@cgps.group', country_value = 'countryX', type_value = 'Epidemiological Metadata' ) [1] "anthony.underwood@cgps.group" [1] "anthony.underwood@cgps.group" [1] "anthony.underwood@cgps.group" qc_and_assembly_metadata <- get_data_for_country('https://docs.google.com/spreadsheets/d/1D04XRov74Cw9LHgZTgkb4s8w4SfqbOOsjwD1k_UdQ-c', user_email = 'anthony.underwood@cgps.group', country_value = 'countryX', type_value = 'QC and Assembly Metadata' ) [1] "anthony.underwood@cgps.group"Now the data can be joined and analysed
Example analysis
In this example we will investiagte the relationship between species ID by Vitek and the genomic prediction using Bactinspector. This involves getting the Epidemiolgical and QC and Assembly metadata first. These dataframes can then be joined and filtered to get counts of where the Vitek matches the genomic resulst (TP) or where they don’t (FP). This can then be plotted as a bar chart and based on this visualisation further investigation performed.
library(tidyr)
library(dplyr)
library(ggplot2)
# select only species data from both tables
epidemiological_metadata_species <- epidemiological_metadata %>% select('Sample id', 'Species')
qc_and_assembly_metadata_species <- qc_and_assembly_metadata %>% select('Sample id', 'Bactinspector species', 'Bactinspector result')
# join epi and qc data
combined_data <- dplyr::inner_join(epidemiological_metadata_species, qc_and_assembly_metadata_species, by='Sample id')
# rename species to epi_three_letter_code
combined_data <- combined_data %>% rename('epi_three_letter_code' = 'Species')
# get species lookup table for whonet 3 letter codes using ghruR function
species_lookup <- whonet_species_map()
# get species name based on the 3 letter code and rename appropriate columns as epi_species and genomic_species
combined_data <- combined_data %>%
left_join(species_lookup, by = c('epi_three_letter_code' = 'three_letter_code')) %>%
rename('epi_species' = 'name', 'genomic_species' = 'Bactinspector species', 'genomic_result_status' = 'Bactinspector result') %>%
select('Sample id', epi_species, genomic_species, genomic_result_status)
# transform salmonella serovars to names matching whonet map
combined_data$genomic_species <- combined_data$genomic_species %>%
stringr::str_replace_all(pattern='Salmonella enterica subsp. enterica serovar (.+)', replacement='Salmonella \\1')
# get true and false positive having removed uncertain genomic_species results
true_and_false_positives <- combined_data %>%
filter(genomic_result_status != 'uncertain') %>%
mutate(match = case_when(genomic_species == epi_species ~ 'TP', TRUE ~ 'FP')) %>%
select(-genomic_species, -genomic_result_status)
# count true and false positives
grouped_data <- true_and_false_positives %>% group_by(epi_species, match) %>% count(name = 'count')
# fill in missing TP or FP with 0
grouped_data <- grouped_data %>% complete(epi_species, match=c('TP', 'FP'), fill = list(count=0))
# plot data
ggplot(grouped_data, aes(fill=match, x = epi_species, y = count)) + geom_bar(position="dodge", stat="identity") + theme(axis.text.x = element_text(angle = 90, hjust = 1))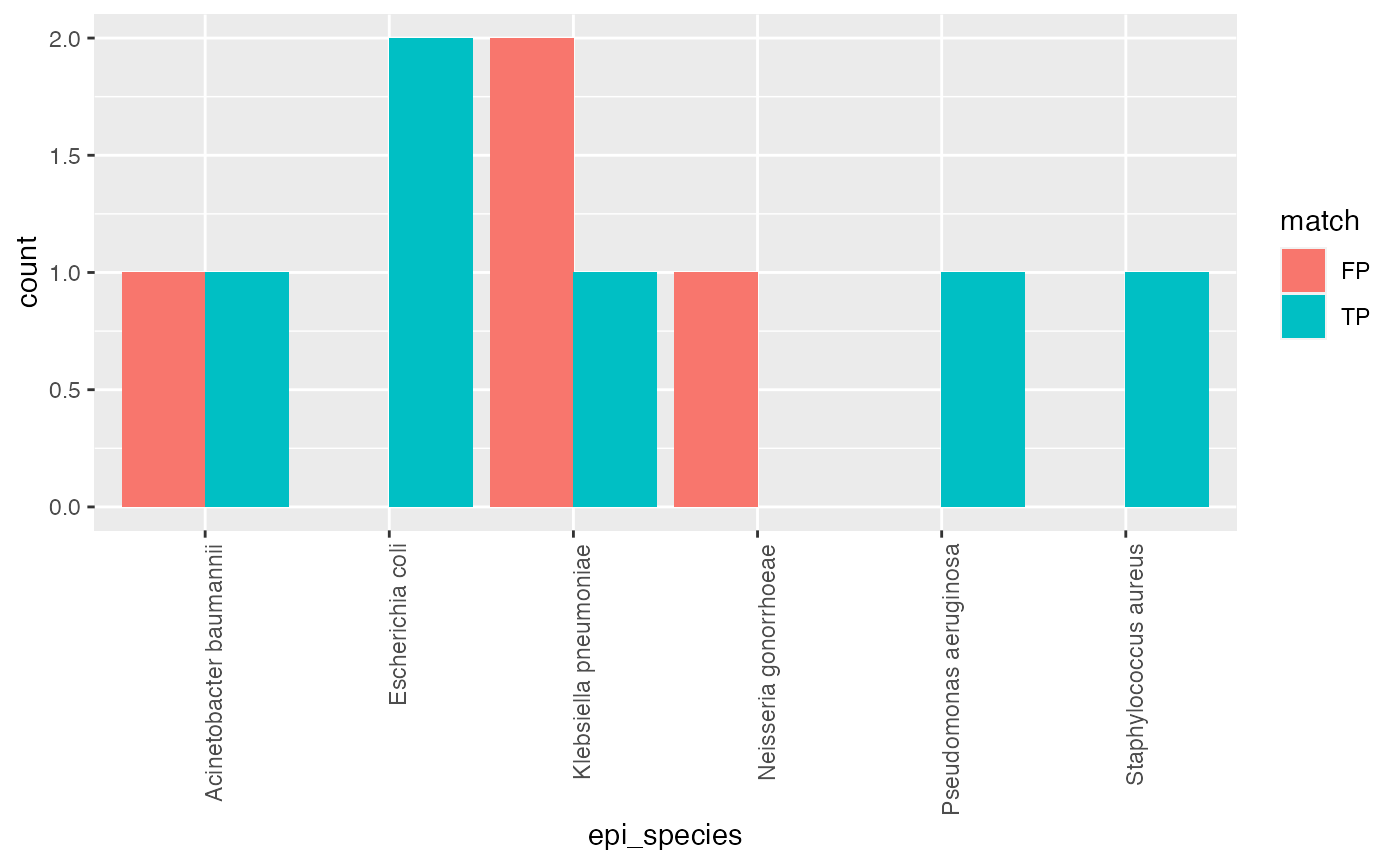
Here is the same plot with some real world data 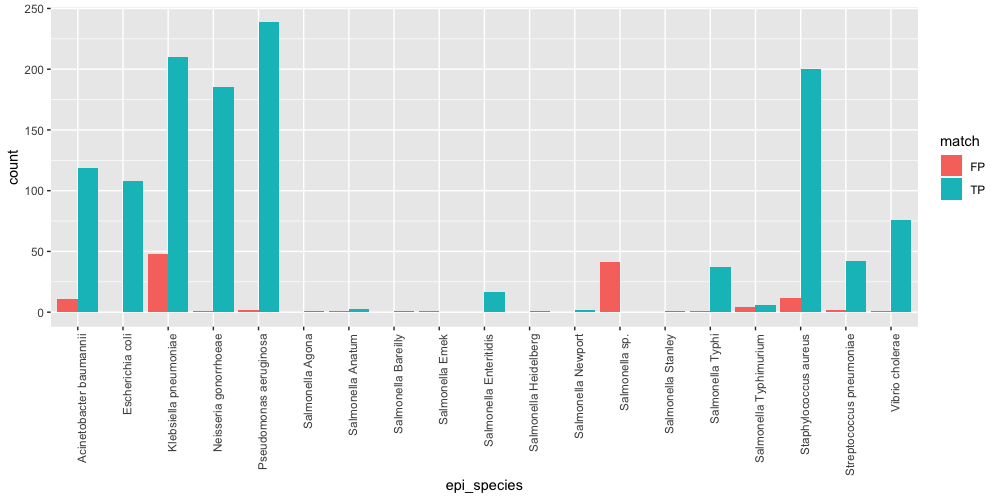 It is interesting to note that Acinetobacter baumannii, Klebsiella pneumoniae, Salmonella, and Staphylococcus aureus have a large number of FP. The Salmonella mismatches are due to issues with genomics prediction of serovar using just mash. The others can be investigated using commands such as
It is interesting to note that Acinetobacter baumannii, Klebsiella pneumoniae, Salmonella, and Staphylococcus aureus have a large number of FP. The Salmonella mismatches are due to issues with genomics prediction of serovar using just mash. The others can be investigated using commands such as
Acinetobacter baumannii
combined_data %>% filter(epi_species == 'Acinetobacter baumannii' & epi_species != genomic_species)
# A tibble: 11 x 4
`Sample id` epi_species genomic_species genomic_result_status
<chr> <chr> <chr> <chr>
1 G18760433 Acinetobacter baumannii Acinetobacter pittii good
2 G18760823 Acinetobacter baumannii Acinetobacter nosocomialis good
3 G18760959 Acinetobacter baumannii Acinetobacter pittii good
4 G18761981 Acinetobacter baumannii Pseudomonas aeruginosa good
5 G18763157 Acinetobacter baumannii Acinetobacter pittii good
6 G18763377 Acinetobacter baumannii Bacillus cereus good
7 G18764158 Acinetobacter baumannii Acinetobacter pittii good
8 G18754211 Acinetobacter baumannii Acinetobacter nosocomialis good
9 G18754214 Acinetobacter baumannii Acinetobacter nosocomialis good
10 G18754246 Acinetobacter baumannii Acinetobacter nosocomialis good
11 G18754295 Acinetobacter baumannii Acinetobacter pittii good Klebsiella pneumoniae
combined_data %>% filter(epi_species == 'Klebsiella pneumoniae' & epi_species != genomic_species)
# A tibble: 48 x 4
`Sample id` epi_species genomic_species genomic_result_status
<chr> <chr> <chr> <chr>
1 G18760420 Klebsiella pneumoniae Klebsiella quasipneumoniae good
2 G18760455 Klebsiella pneumoniae Klebsiella quasipneumoniae good
3 G18760538 Klebsiella pneumoniae Klebsiella quasipneumoniae good
4 G18760678 Klebsiella pneumoniae Klebsiella quasipneumoniae good
5 G18760772 Klebsiella pneumoniae Klebsiella quasipneumoniae good
6 G18761172 Klebsiella pneumoniae Klebsiella quasipneumoniae good
7 G18761185 Klebsiella pneumoniae Klebsiella quasipneumoniae good
8 G18761429 Klebsiella pneumoniae Klebsiella quasipneumoniae good
9 G18762493 Klebsiella pneumoniae Klebsiella quasipneumoniae good
10 G18762494 Klebsiella pneumoniae Klebsiella quasipneumoniae good
# … with 38 more rowsStaphylococcus aureus
combined_data %>% filter(epi_species == 'Staphylococcus aureus' & epi_species != genomic_species)
# A tibble: 13 x 4
`Sample id` epi_species genomic_species genomic_result_status
<chr> <chr> <chr> <chr>
1 G18760291 Staphylococcus aureus Staphylococcus sciuri uncertain
2 G18760340 Staphylococcus aureus Staphylococcus argenteus good
3 G18760797 Staphylococcus aureus Staphylococcus argenteus good
4 G18761989 Staphylococcus aureus Staphylococcus argenteus good
5 G18762363 Staphylococcus aureus Staphylococcus argenteus good
6 G18762364 Staphylococcus aureus Staphylococcus argenteus good
7 G18762634 Staphylococcus aureus Staphylococcus argenteus good
8 G18763441 Staphylococcus aureus Staphylococcus argenteus good
9 G18754525 Staphylococcus aureus Staphylococcus argenteus good
10 G18754612 Staphylococcus aureus Staphylococcus argenteus good
11 G18754695 Staphylococcus aureus Staphylococcus argenteus good
12 G18754745 Staphylococcus aureus Staphylococcus argenteus good
13 G18754754 Staphylococcus aureus Staphylococcus argenteus good