Partitioning gemomic variation
Che-Wei Chang
2021-10-05
Last updated: 2022-01-18
Checks: 6 1
Knit directory: Barley1kGBS_Proj/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20210831) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
- unnamed-chunk-29
- unnamed-chunk-30
- unnamed-chunk-31
- unnamed-chunk-32
- unnamed-chunk-33
- unnamed-chunk-34
To ensure reproducibility of the results, delete the cache directory GenomicVarPartitioning_cache and re-run the analysis. To have workflowr automatically delete the cache directory prior to building the file, set delete_cache = TRUE when running wflow_build() or wflow_publish().
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version da73ff9. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/GenomeEnvAssociation_cache/
Ignored: analysis/GenomicVarPartitioning_cache/
Ignored: analysis/PreprocessEnvData_cache/
Untracked files:
Untracked: VennDiagram2022-01-18_16-08-05.log
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/GenomicVarPartitioning.Rmd) and HTML (public/GenomicVarPartitioning.html) files. If you've configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 5f53139 | Che-Wei Chang | 2022-01-18 | Build site. |
| html | a85e611 | Che-Wei Chang | 2021-11-30 | Build site. |
| Rmd | e201877 | Che-Wei Chang | 2021-10-22 | save changes |
| html | bc726c2 | Che-Wei Chang | 2021-10-22 | Build site. |
| html | 5f82e26 | Che-Wei Chang | 2021-10-22 | Build site. |
| html | f1ac360 | Che-Wei Chang | 2021-10-22 | Build site. |
| html | 1202876 | Che-Wei Chang | 2021-10-22 | Host with GitLab. |
| Rmd | 7c6b41a | Che-Wei Chang | 2021-10-22 | wflow_publish(list.files("./analysis", pattern = "*.Rmd", full.names = T)) |
Introduction
This Rmd displays how to evaluate the genomic variation attributing to environmental gradients, population structure, spatial autocorrelation with the redundancy analysis (RDA). If you use this approach to analyze your data, please cite Lasky et al. (2012).
(The analyses shown here were actually carried in another R script previously. I do not intend to run permutation tests again as they will take hours to finish! Thus, I just copied and pasted R codes and changed the directories to the data. For the demonstration of results, I'll just import the outputs of the previous analyses.)
Load data
rm(list = ls())
source("./code/Rfunctions.R")
library(vcfR)
library(vegan)
# load genotypic data (244 accessions with 27,147 SNPs imputed by Beagle 5 with defaults)
geno <- read.vcfR("./data/GBS_244accessions_27147SNP_Beagle5Imp.vcf.gz", verbose = F)
Y <- t(vcf_to_nummatrix(geno))
colnames(Y) <- geno@fix[,"ID"]
CHR <- as.numeric(geno@fix[,"CHROM"])
POS <- as.numeric(geno@fix[,"POS"])
# load environmental data (244 accessions with 12 environmental variables)
env <- read.delim("./data/Env_data_244accessions_12Env_variables.tsv", header = T)
predictor <- as.data.frame(scale(env)) # standardize data
# load ancestry coefficients (K=4) estimated with ALStructure (https://doi.org/10.1534/genetics.119.302159)
alsQ <- t(readRDS("./data/Ancestry_coeff_244accessions_ALStructure_K4.RDS")$Q_hat)
### double check the sample order
all(mapply(a = rownames(Y), b = gsub(rownames(alsQ), pattern = "HOH-", replacement = ""), function(a,b){grepl(a, pattern = b)})) # all correct[1] TRUEall(rownames(predictor) == rownames(alsQ)) # all correct[1] TRUEMoran's eigenvector maps (MEMs)
We use MEMs to model the spatial autocorrelation in RDA.
MEMs are computed based on the approach of Forester et al. (2018). Please cite their work and also Dray et al. (2006) if you use the R codes showing here.
Please note the forward selection requires considerable computational resources, so it's better to do it with a powerful computer.
library(adespatial)
library(geosphere)
library(spdep)
## load geographic coordinates
geo <- read.table("./data/geo_coordinates_244accessions.tsv", header = T, stringsAsFactors = F)
## calculate mean geographic coordinates of samples from each deme
grp.coord <- cbind(tapply(geo$Longitude, INDEX = geo$GeoGroup, mean), tapply(geo$Latitude, INDEX = geo$GeoGroup, mean))
## 1. generate MEMs
## 1.1 make neighborhood network
nb <- graph2nb(gabrielneigh(grp.coord, nnmult = 100), sym=TRUE) # Gabriel graph: neighbor definition
## 1.2 produce spatial weights matrix
listW <- nb2listw(nb,style="W")
## 1.3 Calculate the distances between neighbours -> 'longlat = TRUE' for longitude-latitude decimal degrees data
disttri <- nbdists(nb,grp.coord, longlat = TRUE)
## 1.4 Use inverse distance as weights
fdist <- lapply(disttri,function(x) x^(-1))
## 1.5 Revised spatial weights matrix
listW <- nb2listw(nb,glist=fdist,style="W")
## 1.6 Eigen analysis of W
MEM <- scores.listw(listW, MEM.autocor = "all")
rownames(MEM) <- rownames(grp.coord)
## 1.7 Match MEMs of demes to samples
mem.full <- as.data.frame(apply(MEM, 2, function(x){x[match(geo$GeoGroup,rownames(MEM))]}))
rownames(mem.full) <- geo$vcfID
## 2. Forward selection
fwsel.mem <- forward.sel(Y = as.matrix(Y), X = as.matrix(mem.full)) # significant if p-value < 0.05
# extract the significant MEMs (p < 0.05)
sel.mem <- mem.full[,colnames(mem.full) %in% fwsel.mem$variables]Partitioning SNP variation
Simple RDA
Contribution of population structure
Ancestry coefficients (K=4) are used to model population structure in the model.
Fit an RDA model
rda.snp.alsQ <- rda(as.matrix(Y) ~ as.matrix(alsQ))Permutation test
rda.snp.alsQ.anova <- anova(rda.snp.alsQ, permutations = 5000)Contribution of spatial autocorrelation
Fit an RDA model
rda.snp.sp <- rda(as.matrix(Y) ~ as.matrix(sel.mem))Permutation test
rda.snp.sp.anova <- anova(rda.snp.sp, permutations = 5000)Contribution of all environmental variables
Fit an RDA model
Here, we fit all environmental variables in an RDA model to evaluate the proportion of genomic variation explained by environments. An ANOVA is performed by the permutation test with the anova function.
rda.snp.env <- rda(as.formula(paste("Y ~ ",paste(colnames(predictor), collapse = " + "), sep = "")), data = predictor)Permutation test
One should notice that permutation will take some time to finish.
rda.snp.env.anova <- anova(rda.snp.env, permutations = 5000)Individual effect of each environmental variable
To evaluate the effect for each environmental variable, we fit one environmental variable in an RDA model at a time. Permutation tests are carried out to test the significance of each environmental variable
env.anova <- lapply(1:ncol(predictor),function(x){anova(rda(Y ~ predictor[,x]), permutations = 5000)} )
names(env.anova) <- colnames(predictor)
# sort the ANOVA result
envstat <- cbind(sapply(env.anova, function(x){x$F[1]}),
sapply(env.anova, function(x){x$Variance[1]}),
sapply(env.anova, function(x){x$Variance[1]/sum(x$Variance)})*100,
sapply(env.anova, function(x){x$`Pr(>F)`[1]}))
rownames(envstat) <- colnames(predictor)
colnames(envstat) <- c("F","Var","PerceVar", "pvalue")
rda.snp.env.anova.ind <- envstat[order(envstat[,1],decreasing = T),]Marginal effect of each environmental variable
By using the argument by = "margin", an environmental variable will be tested with a model given the remnant variables.
rda.snp.env.anova.bymar <- anova.cca(rda.snp.env, model = "direct",by = "margin", parallel = 2,permutations = how(nperm = 5000))Partial RDA controlling population structure
Total effect of all environmental variables
Fit a partial RDA model
rda.snp.env.alsQ <- rda(Y ~ as.matrix(predictor) + Condition(as.matrix(alsQ)), scale = F)Permutation test
rda.snp.env.alsQ.anova <- anova(rda.snp.env.alsQ, permutations = 5000)Individual effect of each environmental variable
env.anova <- lapply(1:ncol(predictor),function(x){anova(rda(Y ~ predictor[,x] + Condition(as.matrix(alsQ))), permutations = 5000)} )
names(env.anova) <- colnames(predictor)
tol.var <- sum(env.anova[[1]]$Variance)
envstat <- cbind(sapply(env.anova, function(x){x$F[1]}),
sapply(env.anova, function(x){x$Variance[1]}),
sapply(env.anova, function(x){x$Variance[1]/tol.var})*100,
sapply(env.anova, function(x){x$`Pr(>F)`[1]}))
rownames(envstat) <- colnames(predictor)
colnames(envstat) <- c("F","Var","PerceVar", "pvalue")
rda.snp.env.alsQ.anova.ind <- envstat[order(envstat[,1],decreasing = T),]Marginal effect of each environmental variable
rda.snp.env.alsQ.anova.bymar <- anova.cca(rda.snp.env.alsQ, model = "direct",by = "margin", parallel = 4, permutations = how(nperm = 5000))Partial RDA controlling spatial autocorrelation
Total effect of all environmental variables
Fit a partial RDA model
rda.snp.env.sp <- rda(as.matrix(Y) ~ as.matrix(predictor) + Condition(as.matrix(sel.mem)))Permutation test
rda.snp.env.sp.anova <- anova(rda.snp.env.sp, permutations = 5000)Partitioning population structure
As the genetic clusters correspond to their eco-geographic regions, it is also interesting to understand the relative contribution of environmental gradients and spatial autocorrelation.
To do so, we take ancestry coefficients as response variables and treat environmental variables and MEMs as explanatory variables in RDA models.
Simple RDA
Contribution of environmental variables
Fit a model
rda.alsQenv <- rda(as.matrix(scale(alsQ)) ~ as.matrix(predictor) )Permutation test
rda.alsQenv.anova <- anova(rda.alsQenv, permutations = 5000)Contribution of spatial autocorrelation
Fit a model
rda.alsQsp <- rda(as.matrix(scale(alsQ)) ~ as.matrix(sel.mem))Permutation test
rda.alsQsp.anova <- anova(rda.alsQsp, permutations = 5000)Partial RDA
Contribution of envieonmental variables
Fit a model controlling spatial autocorrelation
rda.alsQenv.sp <- rda(as.matrix(scale(alsQ)) ~ as.matrix(predictor) + Condition(as.matrix(sel.mem)) )Permutation test
rda.alsQenv.sp.anova <- anova.cca(rda.alsQenv.sp, permutations = 5000)Contribution of spatial autocorrelation
Fit a model controlling environmental variables
rda.alsQsp.env <- rda(as.matrix(scale(alsQ)) ~ as.matrix(sel.mem) + Condition(as.matrix(predictor)))Permutation test
rda.alsQsp.env.anova <- anova(rda.alsQsp.env, permutations = 5000)Results
Partitioning SNP variation
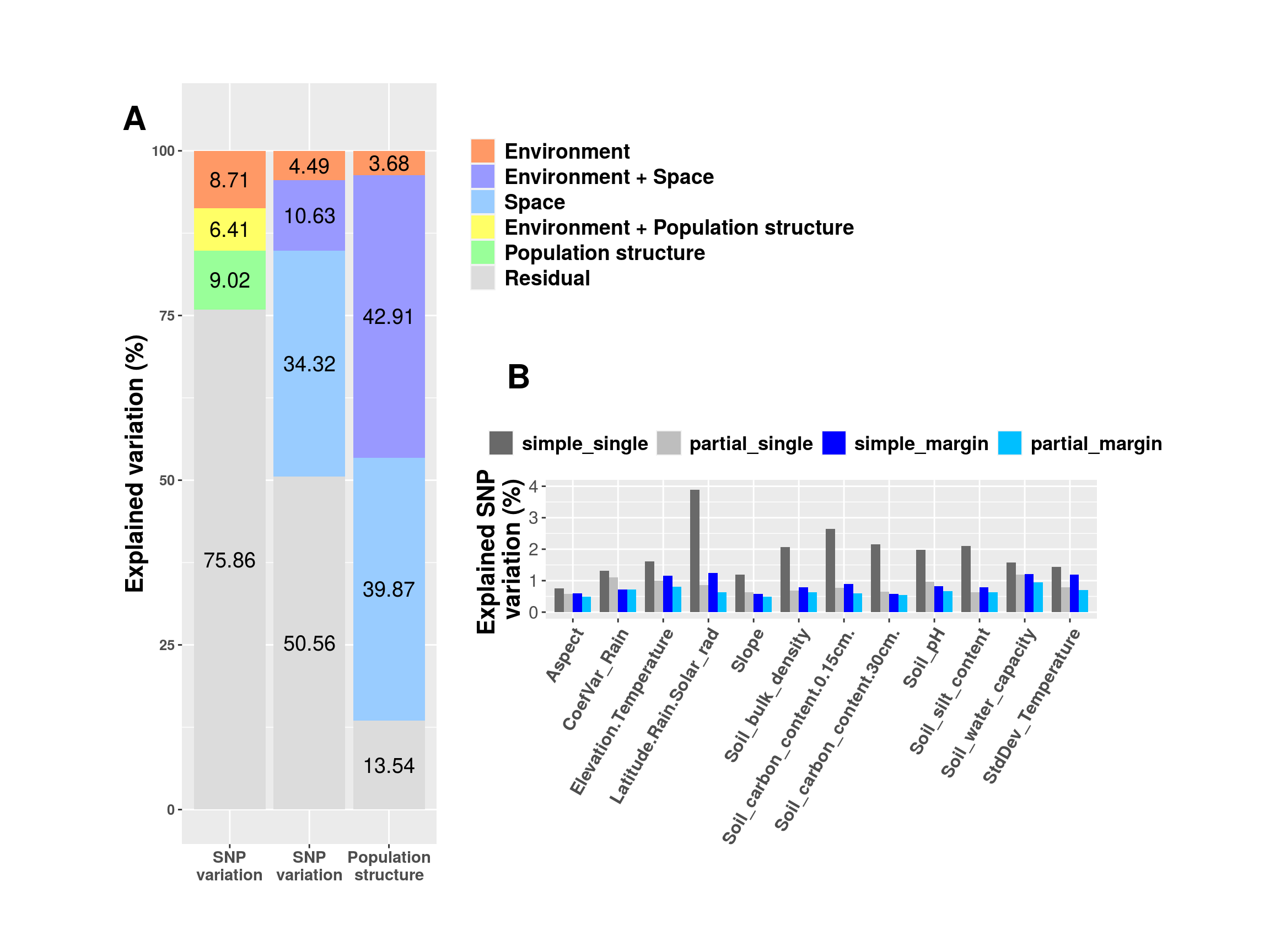
Now, we can look at our results. With the summary table of RDA, we can access the proportion of SNP variation explained by population structure, spatial autocorrelation and environmental variables, respectively. For example, in the summary table of rda.snp.alsQ, we can find that about 15% of variation is explained by the ancestry coefficients (see the column called Proportion).
rda.snp.alsQCall: rda(formula = as.matrix(Y) ~ as.matrix(alsQ))
Inertia Proportion Rank
Total 1.422e+04 1.000e+00
Constrained 2.194e+03 1.543e-01 3
Unconstrained 1.202e+04 8.457e-01 240
Inertia is variance
Some constraints were aliased because they were collinear (redundant)
Eigenvalues for constrained axes:
RDA1 RDA2 RDA3
1017.5 613.7 562.9
Eigenvalues for unconstrained axes:
PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8
370.7 309.5 252.5 199.3 195.6 190.6 174.5 169.7
(Showing 8 of 240 unconstrained eigenvalues)rda.snp.spCall: rda(formula = as.matrix(Y) ~ as.matrix(sel.mem))
Inertia Proportion Rank
Total 1.422e+04 1.000e+00
Constrained 6.391e+03 4.495e-01 56
Unconstrained 7.827e+03 5.505e-01 187
Inertia is variance
Eigenvalues for constrained axes:
RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 RDA7 RDA8 RDA9 RDA10 RDA11 RDA12 RDA13
803.8 574.9 484.6 320.6 215.3 182.1 169.3 164.5 156.8 150.7 135.6 128.2 122.2
RDA14 RDA15 RDA16 RDA17 RDA18 RDA19 RDA20 RDA21 RDA22 RDA23 RDA24 RDA25 RDA26
116.8 110.6 97.8 95.4 92.7 90.9 89.1 87.3 84.8 81.4 79.4 77.0 76.3
RDA27 RDA28 RDA29 RDA30 RDA31 RDA32 RDA33 RDA34 RDA35 RDA36 RDA37 RDA38 RDA39
73.9 71.4 70.3 69.2 68.1 65.9 63.2 61.1 60.1 58.9 58.5 56.4 55.6
RDA40 RDA41 RDA42 RDA43 RDA44 RDA45 RDA46 RDA47 RDA48 RDA49 RDA50 RDA51 RDA52
54.5 54.1 53.0 52.8 52.0 52.0 50.7 50.4 49.5 49.0 47.7 46.7 46.4
RDA53 RDA54 RDA55 RDA56
44.8 38.0 21.5 7.6
Eigenvalues for unconstrained axes:
PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8
333.1 222.1 139.5 129.1 117.6 116.0 106.4 104.0
(Showing 8 of 187 unconstrained eigenvalues)rda.snp.envCall: rda(formula = Y ~ Latitude.Rain.Solar_rad + Aspect + Slope +
Elevation.Temperature + Soil_bulk_density + Soil_water_capacity +
Soil_carbon_content.0.15cm. + Soil_carbon_content.30cm. + Soil_pH +
Soil_silt_content + StdDev_Temperature + CoefVar_Rain, data =
predictor)
Inertia Proportion Rank
Total 1.422e+04 1.000e+00
Constrained 2.150e+03 1.512e-01 12
Unconstrained 1.207e+04 8.488e-01 231
Inertia is variance
Eigenvalues for constrained axes:
RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 RDA7 RDA8 RDA9 RDA10 RDA11 RDA12
627.7 352.3 254.0 183.9 156.6 113.4 93.5 90.8 76.9 73.6 67.5 59.3
Eigenvalues for unconstrained axes:
PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8
530.5 466.3 334.8 264.5 237.0 216.1 173.0 160.8
(Showing 8 of 231 unconstrained eigenvalues)Three RDA models are significant in the permutation test.
rda.snp.alsQ.anovaPermutation test for rda under reduced model
Permutation: free
Number of permutations: 5000
Model: rda(formula = as.matrix(Y) ~ as.matrix(alsQ))
Df Variance F Pr(>F)
Model 3 2194.1 14.597 2e-04 ***
Residual 240 12024.5
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1rda.snp.sp.anovaPermutation test for rda under reduced model
Permutation: free
Number of permutations: 5000
Model: rda(formula = as.matrix(Y) ~ as.matrix(sel.mem))
Df Variance F Pr(>F)
Model 56 6391.3 2.7267 2e-04 ***
Residual 187 7827.3
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1rda.snp.env.anovaPermutation test for rda under reduced model
Permutation: free
Number of permutations: 5000
Model: rda(formula = Y ~ Latitude.Rain.Solar_rad + Aspect + Slope + Elevation.Temperature + Soil_bulk_density + Soil_water_capacity + Soil_carbon_content.0.15cm. + Soil_carbon_content.30cm. + Soil_pH + Soil_silt_content + StdDev_Temperature + CoefVar_Rain, data = predictor)
Df Variance F Pr(>F)
Model 12 2149.7 3.4288 2e-04 ***
Residual 231 12068.9
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1To calculate adjusted \(R^2\), we can use RsquareAdj function.
RsquareAdj(rda.snp.alsQ)$r.squared
[1] 0.1543112
$adj.r.squared
[1] 0.1437401RsquareAdj(rda.snp.sp)$r.squared
[1] 0.4495023
$adj.r.squared
[1] 0.2846473RsquareAdj(rda.snp.env)$r.squared
[1] 0.1511914
$adj.r.squared
[1] 0.1070974Similarly, we can access the confounded variation by looking at the summary tables. For example, in the summary table of rda.snp.env.sp, we can find that about 45% of variation is explained by MEMs (see the row Conditional) and only 4.5% of variation is exclusively explained by environmental variables.
rda.snp.env.alsQCall: rda(formula = Y ~ as.matrix(predictor) +
Condition(as.matrix(alsQ)), scale = F)
Inertia Proportion Rank
Total 1.422e+04 1.000e+00
Conditional 2.194e+03 1.543e-01 3
Constrained 1.238e+03 8.710e-02 12
Unconstrained 1.079e+04 7.586e-01 228
Inertia is variance
Some constraints were aliased because they were collinear (redundant)
Eigenvalues for constrained axes:
RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 RDA7 RDA8 RDA9 RDA10 RDA11
204.12 177.36 141.23 119.24 100.27 84.85 82.65 77.01 70.10 66.69 58.73
RDA12
56.19
Eigenvalues for unconstrained axes:
PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8
286.55 240.21 225.75 173.28 161.47 157.15 148.16 141.90
(Showing 8 of 228 unconstrained eigenvalues)rda.snp.env.spCall: rda(formula = as.matrix(Y) ~ as.matrix(predictor) +
Condition(as.matrix(sel.mem)))
Inertia Proportion Rank
Total 1.422e+04 1.000e+00
Conditional 6.391e+03 4.495e-01 56
Constrained 6.386e+02 4.491e-02 12
Unconstrained 7.189e+03 5.056e-01 175
Inertia is variance
Eigenvalues for constrained axes:
RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 RDA7 RDA8 RDA9 RDA10 RDA11
135.57 72.64 59.91 50.45 48.26 45.74 41.74 40.52 38.01 36.81 34.58
RDA12
34.34
Eigenvalues for unconstrained axes:
PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8
274.41 195.45 135.70 117.92 114.12 104.91 103.22 100.38
(Showing 8 of 175 unconstrained eigenvalues)RsquareAdj(rda.snp.env.alsQ)$r.squared
[1] 0.08709915
$adj.r.squared
[1] 0.04776304RsquareAdj(rda.snp.env.sp)$r.squared
[1] 0.04491004
$adj.r.squared
[1] 0.01330805rda.snp.env.alsQ.anovaPermutation test for rda under reduced model
Permutation: free
Number of permutations: 5000
Model: rda(formula = Y ~ as.matrix(predictor) + Condition(as.matrix(alsQ)), scale = F)
Df Variance F Pr(>F)
Model 12 1238.4 2.1815 2e-04 ***
Residual 228 10786.1
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1rda.snp.env.sp.anovaPermutation test for rda under reduced model
Permutation: free
Number of permutations: 5000
Model: rda(formula = as.matrix(Y) ~ as.matrix(predictor) + Condition(as.matrix(sel.mem)))
Df Variance F Pr(>F)
Model 12 638.6 1.2954 2e-04 ***
Residual 175 7188.8
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1We can further investigate the individual effects and marginal effects of environmental variables.
# make tables summarizing the anova results for all environmental variables (one variable was fitted in an RDA model at a time)
env.names <- names(rda.snp.env.anova.ind)
rda.snp.env.anova.ind <- round(matrix(unlist(lapply(rda.snp.env.anova.ind, function(x){c(x$Df[1], x$Variance[1], x$F[1], x$"Pr(>F)"[1])})), ncol = 4, byrow = T), digits = 4)
rda.snp.env.alsQ.anova.ind <- round(matrix(unlist(lapply(rda.snp.env.alsQ.anova.ind, function(x){c(x$Df[1], x$Variance[1], x$F[1], x$"Pr(>F)"[1])})), ncol = 4, byrow = T), digits = 4)
# name the rows and columns
colnames(rda.snp.env.anova.ind) <- colnames(rda.snp.env.alsQ.anova.ind) <- c("Df", "Variance", "F", "Pr(>F)")
rownames(rda.snp.env.anova.ind) <- rownames(rda.snp.env.alsQ.anova.ind) <- env.names
# individual effects and marginal effects without conditioning on population structure
rda.snp.env.anova.ind Df Variance F Pr(>F)
Latitude.Rain.Solar_rad 1 553.6372 9.8046 2e-04
Aspect 1 107.0534 1.8359 2e-04
Slope 1 170.2299 2.9324 2e-04
Elevation.Temperature 1 229.2979 3.9666 2e-04
Soil_bulk_density 1 293.9198 5.1081 2e-04
Soil_water_capacity 1 225.4961 3.8998 2e-04
Soil_carbon_content.0.15cm. 1 376.0355 6.5739 2e-04
Soil_carbon_content.30cm. 1 306.0195 5.3230 2e-04
Soil_pH 1 280.3609 4.8677 2e-04
Soil_silt_content 1 298.7987 5.1947 2e-04
StdDev_Temperature 1 204.1527 3.5253 2e-04
CoefVar_Rain 1 186.4980 3.2164 2e-04rda.snp.env.anova.bymarPermutation test for rda under direct model
Marginal effects of terms
Permutation: free
Number of permutations: 5000
Model: rda(formula = Y ~ Latitude.Rain.Solar_rad + Aspect + Slope + Elevation.Temperature + Soil_bulk_density + Soil_water_capacity + Soil_carbon_content.0.15cm. + Soil_carbon_content.30cm. + Soil_pH + Soil_silt_content + StdDev_Temperature + CoefVar_Rain, data = predictor)
Df Variance F Pr(>F)
Latitude.Rain.Solar_rad 1 177.1 3.3906 0.0002000 ***
Aspect 1 85.5 1.6364 0.0017996 **
Slope 1 81.9 1.5674 0.0023995 **
Elevation.Temperature 1 164.8 3.1535 0.0002000 ***
Soil_bulk_density 1 112.7 2.1565 0.0002000 ***
Soil_water_capacity 1 171.6 3.2845 0.0002000 ***
Soil_carbon_content.0.15cm. 1 128.0 2.4493 0.0002000 ***
Soil_carbon_content.30cm. 1 81.4 1.5571 0.0053989 **
Soil_pH 1 117.1 2.2410 0.0002000 ***
Soil_silt_content 1 112.0 2.1446 0.0003999 ***
StdDev_Temperature 1 169.4 3.2423 0.0002000 ***
CoefVar_Rain 1 103.3 1.9776 0.0002000 ***
Residual 231 12068.9
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1# individual effects and marginal effects controlling the effect of population structure
rda.snp.env.alsQ.anova.ind Df Variance F Pr(>F)
Latitude.Rain.Solar_rad 1 122.1676 2.4531 2e-04
Aspect 1 81.2028 1.6250 2e-04
Slope 1 90.5932 1.8143 2e-04
Elevation.Temperature 1 141.3632 2.8432 2e-04
Soil_bulk_density 1 98.1368 1.9666 2e-04
Soil_water_capacity 1 168.3390 3.3934 2e-04
Soil_carbon_content.0.15cm. 1 110.8457 2.2237 2e-04
Soil_carbon_content.30cm. 1 92.3804 1.8504 2e-04
Soil_pH 1 136.2396 2.7389 2e-04
Soil_silt_content 1 89.6590 1.7954 2e-04
StdDev_Temperature 1 111.8356 2.2437 2e-04
CoefVar_Rain 1 156.2026 3.1455 2e-04rda.snp.env.alsQ.anova.bymarPermutation test for rda under direct model
Marginal effects of terms
Permutation: free
Number of permutations: 5000
Model: rda(formula = Y ~ Latitude.Rain.Solar_rad + Aspect + Slope + Elevation.Temperature + Soil_bulk_density + Soil_water_capacity + Soil_carbon_content.0.15cm. + Soil_carbon_content.30cm. + Soil_pH + Soil_silt_content + StdDev_Temperature + CoefVar_Rain + Condition(North) + Condition(Coast) + Condition(Eastern_Desert) + Condition(Southern_Desert), data = cbind.data.frame(predictor, alsQ))
Df Variance F Pr(>F)
Latitude.Rain.Solar_rad 1 89.2 1.8864 0.0002000 ***
Aspect 1 69.9 1.4777 0.0089982 **
Slope 1 70.0 1.4805 0.0079984 **
Elevation.Temperature 1 114.6 2.4234 0.0002000 ***
Soil_bulk_density 1 89.8 1.8989 0.0005999 ***
Soil_water_capacity 1 134.4 2.8405 0.0002000 ***
Soil_carbon_content.0.15cm. 1 85.1 1.7986 0.0005999 ***
Soil_carbon_content.30cm. 1 77.2 1.6314 0.0033993 **
Soil_pH 1 95.4 2.0162 0.0005999 ***
Soil_silt_content 1 88.7 1.8743 0.0003999 ***
StdDev_Temperature 1 100.3 2.1193 0.0002000 ***
CoefVar_Rain 1 101.8 2.1510 0.0002000 ***
Residual 228 10786.1
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Finally, we visualize the results of variation partitioning.
| Version | Author | Date |
|---|---|---|
| 1202876 | Che-Wei Chang | 2021-10-22 |
sessionInfo()R version 3.6.0 (2019-04-26)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 16.04.6 LTS
Matrix products: default
BLAS: /usr/lib/libblas/libblas.so.3.6.0
LAPACK: /usr/lib/lapack/liblapack.so.3.6.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggplot2_3.3.5 vegan_2.5-6 lattice_0.20-38 permute_0.9-5
[5] vcfR_1.9.0 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] tidyselect_1.1.1 xfun_0.27 purrr_0.3.4 memuse_4.0-0
[5] splines_3.6.0 colorspace_2.0-2 vctrs_0.3.8 generics_0.1.0
[9] htmltools_0.5.2 viridisLite_0.4.0 yaml_2.2.1 mgcv_1.8-28
[13] utf8_1.2.2 rlang_0.4.12 later_1.3.0 pillar_1.6.4
[17] withr_2.4.2 DBI_1.1.1 glue_1.4.2 lifecycle_1.0.1
[21] stringr_1.4.0 munsell_0.5.0 gtable_0.3.0 evaluate_0.14
[25] labeling_0.4.2 knitr_1.36 fastmap_1.1.0 httpuv_1.6.3
[29] parallel_3.6.0 fansi_0.5.0 highr_0.9 Rcpp_1.0.7
[33] pinfsc50_1.1.0 promises_1.2.0.1 backports_1.1.5 scales_1.1.1
[37] farver_2.1.0 fs_1.5.0 digest_0.6.28 stringi_1.7.5
[41] dplyr_1.0.7 grid_3.6.0 rprojroot_1.3-2 tools_3.6.0
[45] magrittr_2.0.1 tibble_3.1.5 cluster_2.0.8 crayon_1.4.1
[49] ape_5.3 whisker_0.4 pkgconfig_2.0.3 ellipsis_0.3.2
[53] MASS_7.3-51.1 Matrix_1.2-18 assertthat_0.2.1 rmarkdown_2.3
[57] R6_2.5.1 nlme_3.1-139 git2r_0.26.1 compiler_3.6.0