MS: Identifying gene modules with non-negative matrix factorization (pseudo-bulk)
Will Macnair
Institute for Molecular Life Sciences, University of Zurich, SwitzerlandSwiss Institute of Bioinformatics (SIB), University of Zurich, SwitzerlandSeptember 28, 2021
Last updated: 2021-09-28
Checks: 4 3
Knit directory: MS_lesions/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
The global environment had objects present when the code in the R Markdown file was run. These objects can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment. Use wflow_publish or wflow_build to ensure that the code is always run in an empty environment.
The following objects were defined in the global environment when these results were created:
| Name | Class | Size |
|---|---|---|
| q | function | 1008 bytes |
The command set.seed(20210118) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
- load_fine_expression
- load_inputs
- load_popalign_outputs
- load_soup
- load_umap
- plot_annotations_more
- plot_annotations_std
- plot_celltypes_over_umap
- plot_genes_dotplot
- plot_genes
- plot_genes_heatmap
- plot_scores_by_type_scaled_2
- plot_scores_by_type_scaled
- plot_scores_by_type_unscaled
- plot_scores_over_lesions
- plot_umap_celltypes
- run_onmf
- save_outputs
- save_outputs_for_popalign
- save_sub_sces
- session_info
- session-info-chunk-inserted-by-workflowr
- setup_input
- setup_outputs
To ensure reproducibility of the results, delete the cache directory ms08_modules_pseudobulk_cache and re-run the analysis. To have workflowr automatically delete the cache directory prior to building the file, set delete_cache = TRUE when running wflow_build() or wflow_publish().
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 62b9816. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rprofile
Ignored: .Rproj.user/
Ignored: ._MS_lesions.sublime-project
Ignored: .log/
Ignored: MS_lesions.sublime-project
Ignored: MS_lesions.sublime-workspace
Ignored: analysis/.__site.yml
Ignored: analysis/fig_muscat_cache/
Ignored: analysis/ms02_doublet_id_cache/
Ignored: analysis/ms03_SampleQC_cache/
Ignored: analysis/ms04_conos_cache/
Ignored: analysis/ms05_splitting_cache/
Ignored: analysis/ms06_sccaf_cache/
Ignored: analysis/ms07_soup_cache/
Ignored: analysis/ms08_modules_cache/
Ignored: analysis/ms08_modules_pseudobulk_cache/
Ignored: analysis/ms09_ancombc_cache/
Ignored: analysis/ms09_ancombc_clean_1e3_cache/
Ignored: analysis/ms09_ancombc_clean_2e3_cache/
Ignored: analysis/ms10_muscat_run01_cache/
Ignored: analysis/ms10_muscat_run02_cache/
Ignored: analysis/ms10_muscat_template_broad_cache/
Ignored: analysis/ms10_muscat_template_fine_cache/
Ignored: analysis/ms11_paga_cache/
Ignored: analysis/ms12_markers_cache/
Ignored: analysis/ms13_labelling_cache/
Ignored: analysis/ms14_lesions_cache/
Ignored: analysis/ms15_mofa_sample_gm_cache/
Ignored: analysis/ms15_mofa_sample_gm_superclean_cache/
Ignored: analysis/ms15_mofa_sample_wm_cache/
Ignored: analysis/ms15_mofa_sample_wm_new_meta_cache/
Ignored: analysis/ms15_mofa_sample_wm_superclean_cache/
Ignored: analysis/ms15_patients_cache/
Ignored: analysis/ms15_patients_gm_cache/
Ignored: analysis/ms15_patients_sample_level_cache/
Ignored: analysis/ms15_patients_w_ms_cache/
Ignored: analysis/supp06_sccaf_cache/
Ignored: analysis/supp07_superclean_check_cache/
Ignored: analysis/supp09_ancombc_cache/
Ignored: analysis/supp09_ancombc_mixed_cache/
Ignored: analysis/supp10_muscat_cache/
Ignored: analysis/supp10_muscat_ctrl_gm_vs_wm_cache/
Ignored: analysis/supp10_muscat_heatmaps_cache/
Ignored: analysis/supp10_muscat_olg_pc1_cache/
Ignored: analysis/supp10_muscat_olg_pc2_cache/
Ignored: analysis/supp10_muscat_olg_pc_cache/
Ignored: analysis/supp10_muscat_regression_cache/
Ignored: analysis/supp10_muscat_soup_cache/
Ignored: code/.recovery/
Ignored: code/muscat_plan.txt
Ignored: data/
Ignored: figures/
Ignored: output/
Ignored: tmp/
Untracked files:
Untracked: Rplots.pdf
Untracked: analysis/supp09_ancombc_mixed.Rmd
Untracked: code/ancom_v2.1.R
Untracked: code/dev_check_oligo_groupings_20210924.R
Untracked: code/jobs/muscat_job_2021-09-22_broad.slurm
Untracked: code/metadata_update_plan_20210928.txt
Unstaged changes:
Modified: analysis/ms08_modules_pseudobulk.Rmd
Modified: analysis/ms09_ancombc.Rmd
Modified: analysis/ms14_lesions.Rmd
Modified: analysis/ms15_mofa_sample_wm_new_meta.Rmd
Modified: code/dev_check_pmi_distns_20210824.R
Modified: code/dev_stan_cauchy_fits_2021-06-28.R
Modified: code/ms08_modules.R
Modified: code/ms09_ancombc.R
Modified: code/ms10_muscat_fns.R
Modified: code/ms10_muscat_runs.R
Modified: code/ms14_lesions.R
Modified: code/ms15_mofa.R
Modified: code/supp09_ancombc.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ms08_modules_pseudobulk.Rmd) and HTML (docs/ms08_modules_pseudobulk.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 7ddc417 | Macnair | 2021-09-24 | Add pseudobulk version of modules |
| html | 7ddc417 | Macnair | 2021-09-24 | Add pseudobulk version of modules |
Setup / definitions
Libraries
Helper functions
source('code/ms00_utils.R')
source('code/ms08_modules.R')
source_python('code/ms08_modules.py')Inputs
# base inputs
sce_f = 'data/sce_raw/ms_sce.rds'
labels_f = 'data/byhand_markers/validation_markers_2021-05-31.csv'
labelled_f = 'output/ms13_labelling/conos_labelled_2021-05-31.txt.gz'
meta_f = 'data/metadata/metadata_updated_20201127.txt'
# define pseudobulk files
soup_dir = 'output/ms07_soup'
date_soup = '2021-06-01'
pb_fine_f = sprintf('%s/pb_sum_fine_%s.rds', soup_dir, date_soup)
prop_fine_f = sprintf('%s/pb_prop_fine_%s.rds', soup_dir, date_soup)
# broad level pseudobulk files
pb_broad_f = file.path(soup_dir, 'pb_sum_broad_2021-06-01.rds')
pb_fine_f = file.path(soup_dir, 'pb_sum_fine_2021-06-01.rds')
prop_fine_f = file.path(soup_dir, 'pb_prop_fine_2021-06-01.rds')
pb_soup_f = file.path(soup_dir, 'pb_soup_broad_maximum_2021-06-01.rds')
gtf_f = 'data/gtf/Homo_sapiens.GRCh38.96.filtered.preMRNA.gtf'Outputs
# set up directory
save_dir = 'output/ms08_modules'
date_tag = '2021-09-24'
if (!dir.exists(save_dir))
dir.create(save_dir)
ncores = 8
# output file patterns
genes_pat = sprintf('%s/%s/features_%s_%s.tsv', save_dir, '%s', date_tag, '%s')
mtx_pat = sprintf('%s/%s/counts_%s_%s.mtx', save_dir, '%s', date_tag, '%s')
sce_pat = sprintf('%s/%s/pb_sub_%s_%s.rds', save_dir, '%s', date_tag, '%s')
ok_gs_pat = sprintf('%s/%s/ok_gs_%s_%s.txt', save_dir, '%s', date_tag, '%s')
pop_pat = sprintf('%s/%s/pop_%s_%s.p', save_dir, '%s', '%s', date_tag)
res_pat = sprintf('%s/%s/res_%s_%s.rds', save_dir, '%s', date_tag, '%s')
go_pat = sprintf('%s/%s/go_dt_%s_%s_%s.rds', save_dir, '%s', date_tag, '%s', '%s')
# lists of celltypes for each run
spec_list = list(
oligo_opc = list(type_broad = c('OPCs / COPs', 'Oligodendrocytes')),
micro_immune = list(type_broad = c('Microglia', 'Immune')),
excitatory = list(type_broad = 'Excitatory neurons'),
inhibitory = list(type_broad = 'Inhibitory neurons'),
astrocytes = list(type_broad = c('Astrocytes')),
endo_stromal = list(type_broad = c('Endothelial cells', 'Pericytes')),
microglia = list(type_broad = c('Microglia')),
immune = list(type_broad = c('Immune'))
)
assert_that(length(spec_list) == length(unique(names(spec_list))))[1] TRUEgroup_list = names(spec_list)
# how many per fine celltype?
n_sample = 2e3
n_genes = 2e3
max_soup = 0.1
ok_types = 'protein_coding'
# umap params
umap_many_f = 'output/ms04_conos/conos_umap_sub_2021-02-11.txt'
# umap_ps = list(
# min_dist = 0.1,
# spread = 8
# )
umap_ps = list(
min_dist = 1,
spread = 2
)
# define xls file to save
xl_f = sprintf('%s/modules_genes_%s.xlsx', save_dir, date_tag)Load inputs
labels_dt = load_names_dt(labels_f) %>%
.[, cluster_id := type_fine]
conos_dt = load_labelled_dt(labelled_f, labels_f)
meta_dt = load_meta_dt(meta_f)pb_soup = pb_soup_f %>% readRDS
pb_broad = pb_broad_f %>% readRDS
contam_dt = calc_contam_dt(pb_soup, pb_broad, min_cells = 10)
rm(pb_soup, pb_broad)
biotypes_dt = get_biotypes_dt(gtf_f)fine_dt = load_fine_dt(pb_fine_f, prop_fine_f, labels_dt)umap_dt = umap_many_f %>% fread %>%
.[ min_dist == umap_ps$min_dist & spread == umap_ps$spread ] %>%
.[, .(cell_id, UMAP1, UMAP2)]Processing / calculations
save_sub_sces_pb(spec_list, pb_fine_f, sce_pat, ok_gs_pat, labels_dt, contam_dt,
biotypes_dt, n_sample, n_genes, max_soup, ok_types, min_cells = 10, save_dir)already done!NULLsave_outputs_for_popalign(group_list, sce_pat, mtx_pat, genes_pat)already done!NULLrun_onmf(save_dir, date_tag, group_list, ncores = ncores)pop_list = group_list %>%
map(~get_pop_results(.x, res_pat, sce_pat, ok_gs_pat, go_pat, pop_pat,
conos_dt, labels_dt, is_pb = TRUE)) %>% setNames(group_list)Analysis
Median module scores by celltype (scaled)
for (g in group_list) {
cat('### ', g, '\n')
hm = plot_scores_by_celltype(pop_list[[g]]$scores_dt, pop_list[[g]]$k_order,
what = 'scaled')
if (!is.null(hm))
draw(hm)
cat('\n\n')
}oligo_opc
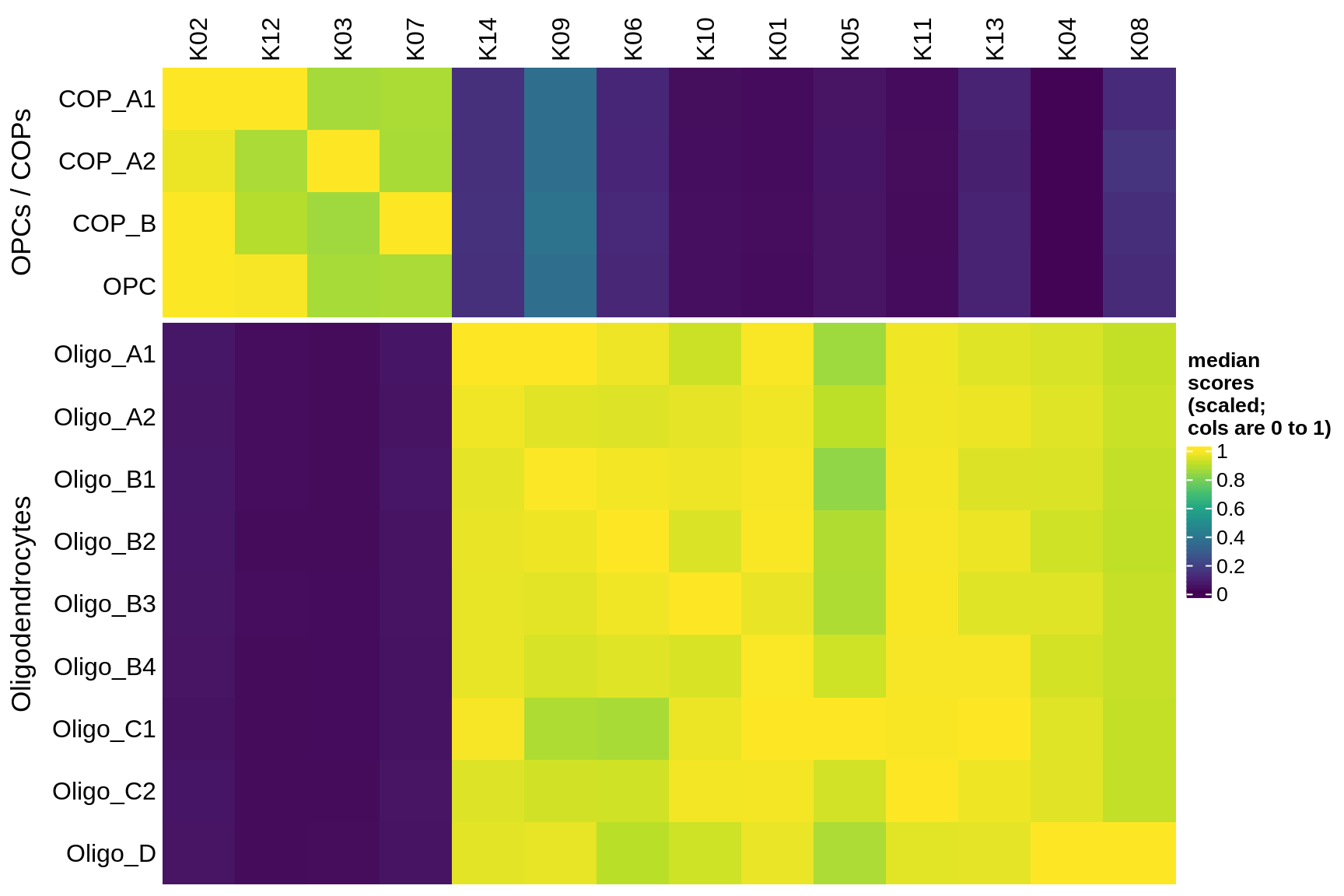
micro_immune
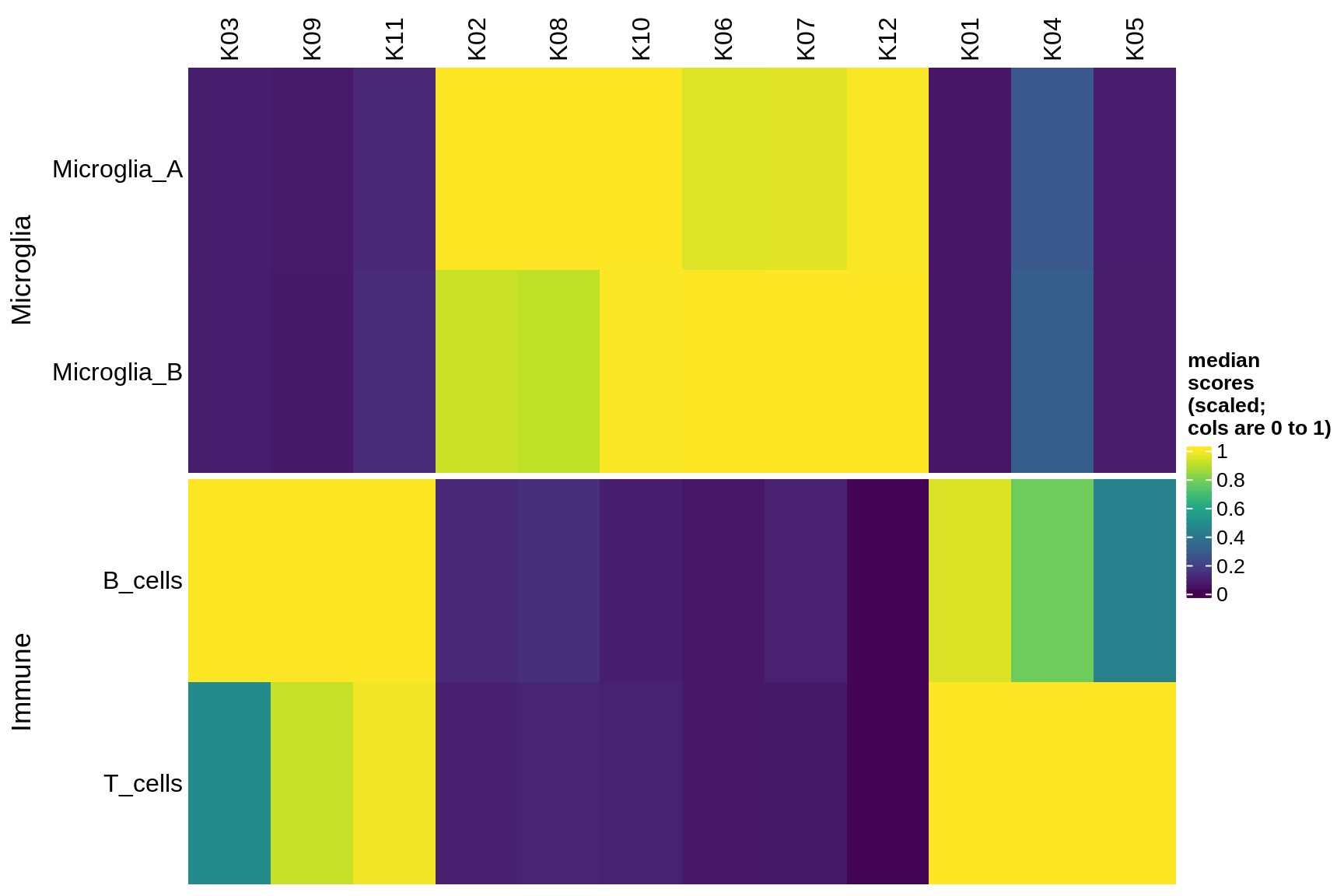
excitatory
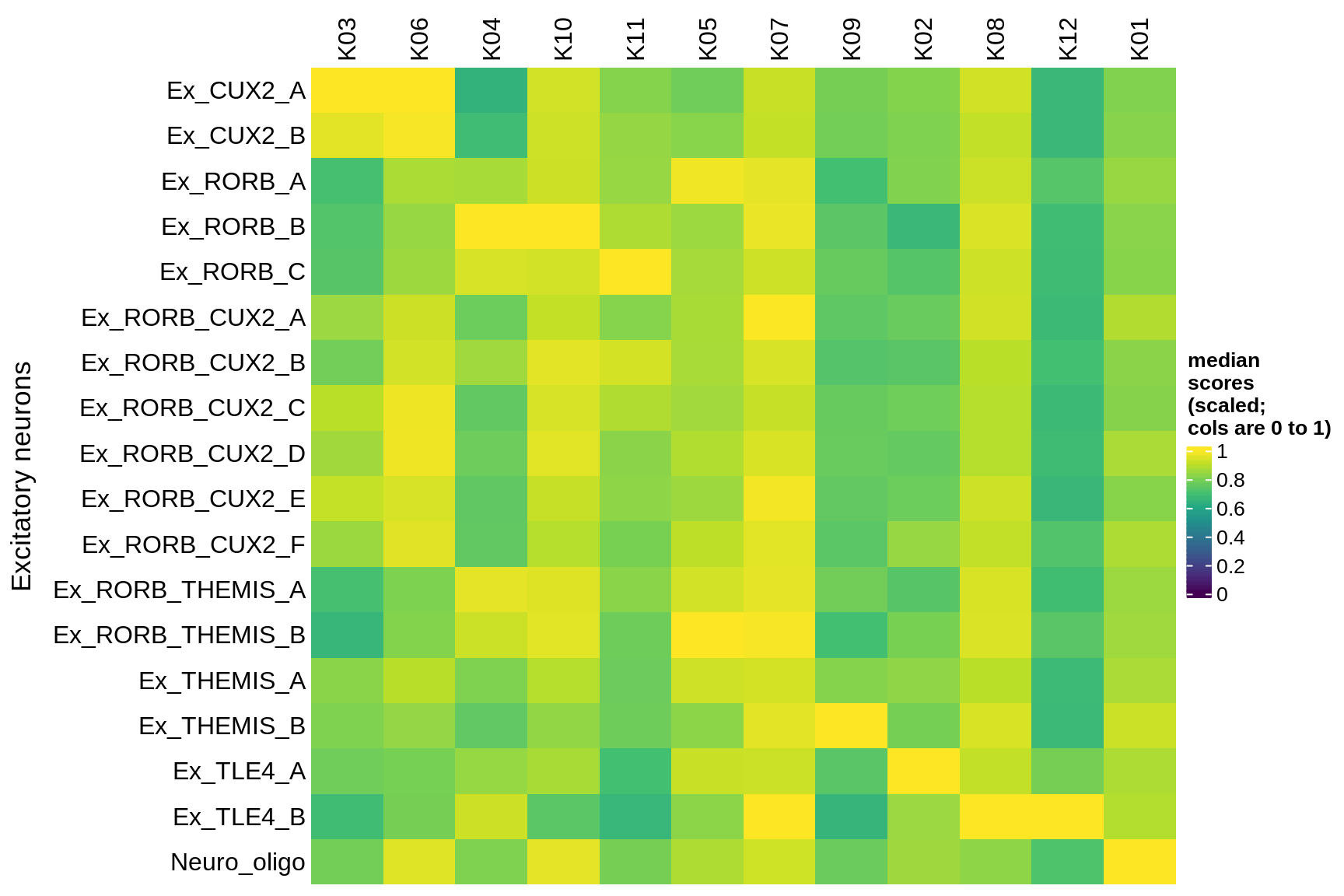
inhibitory
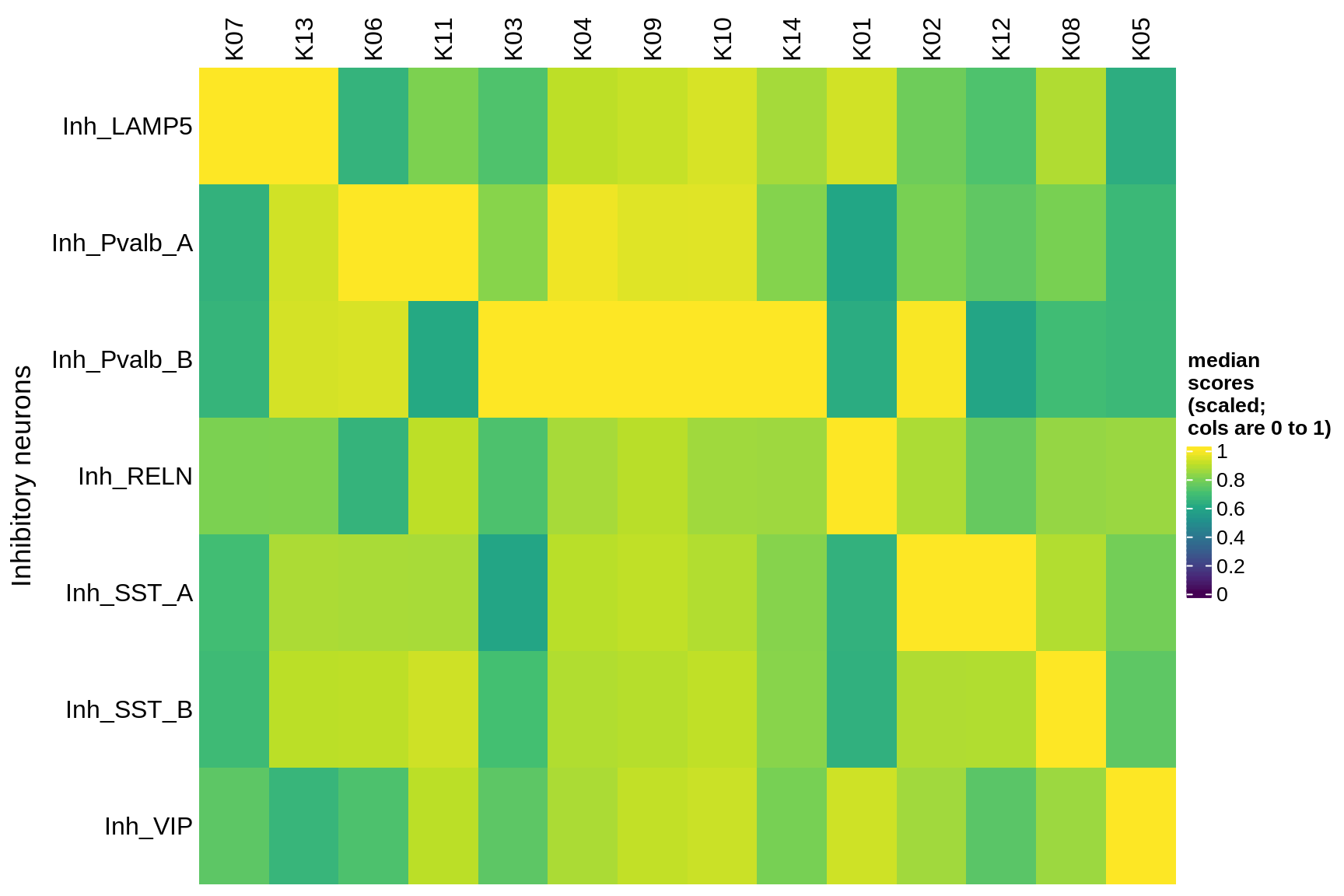
astrocytes
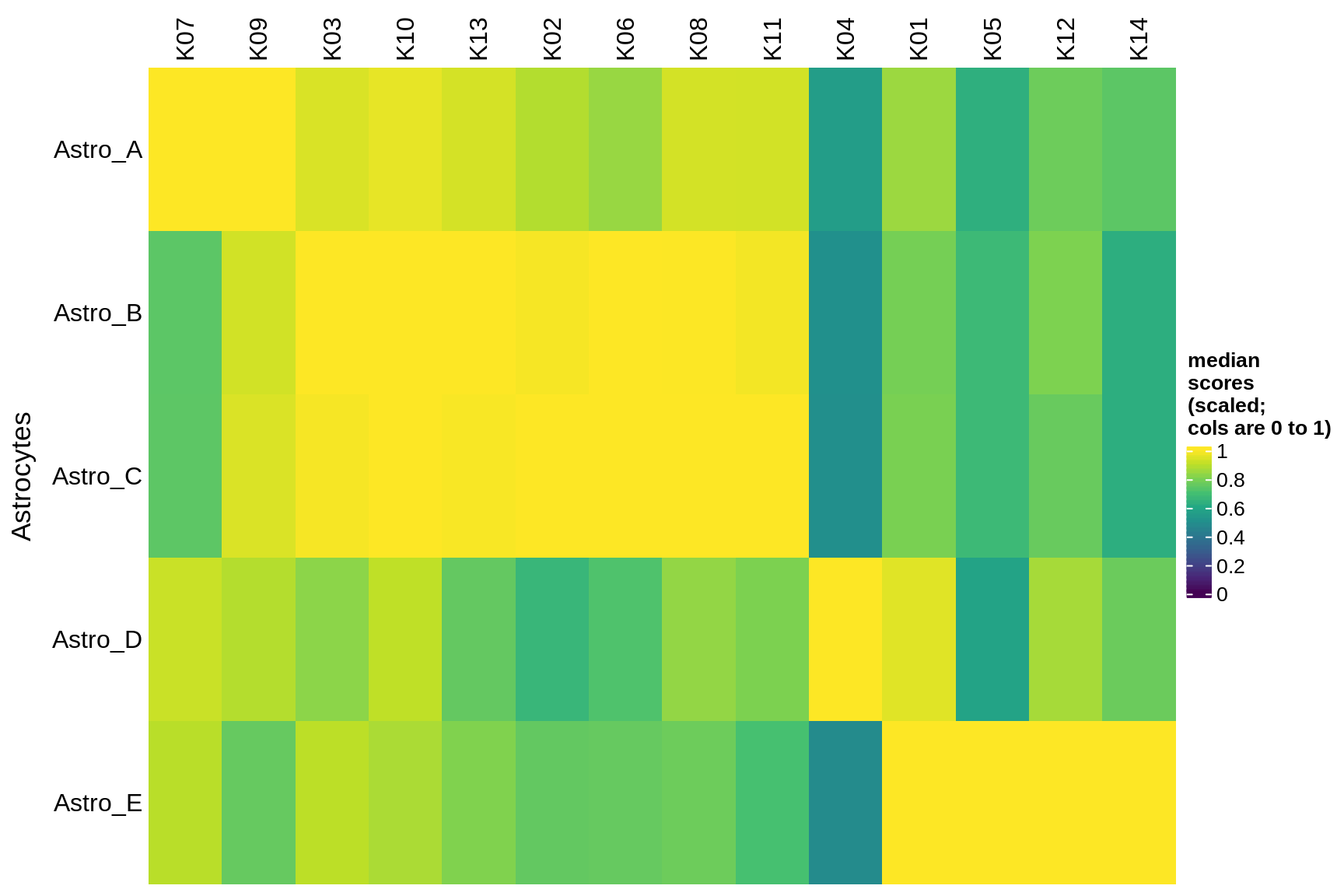
endo_stromal
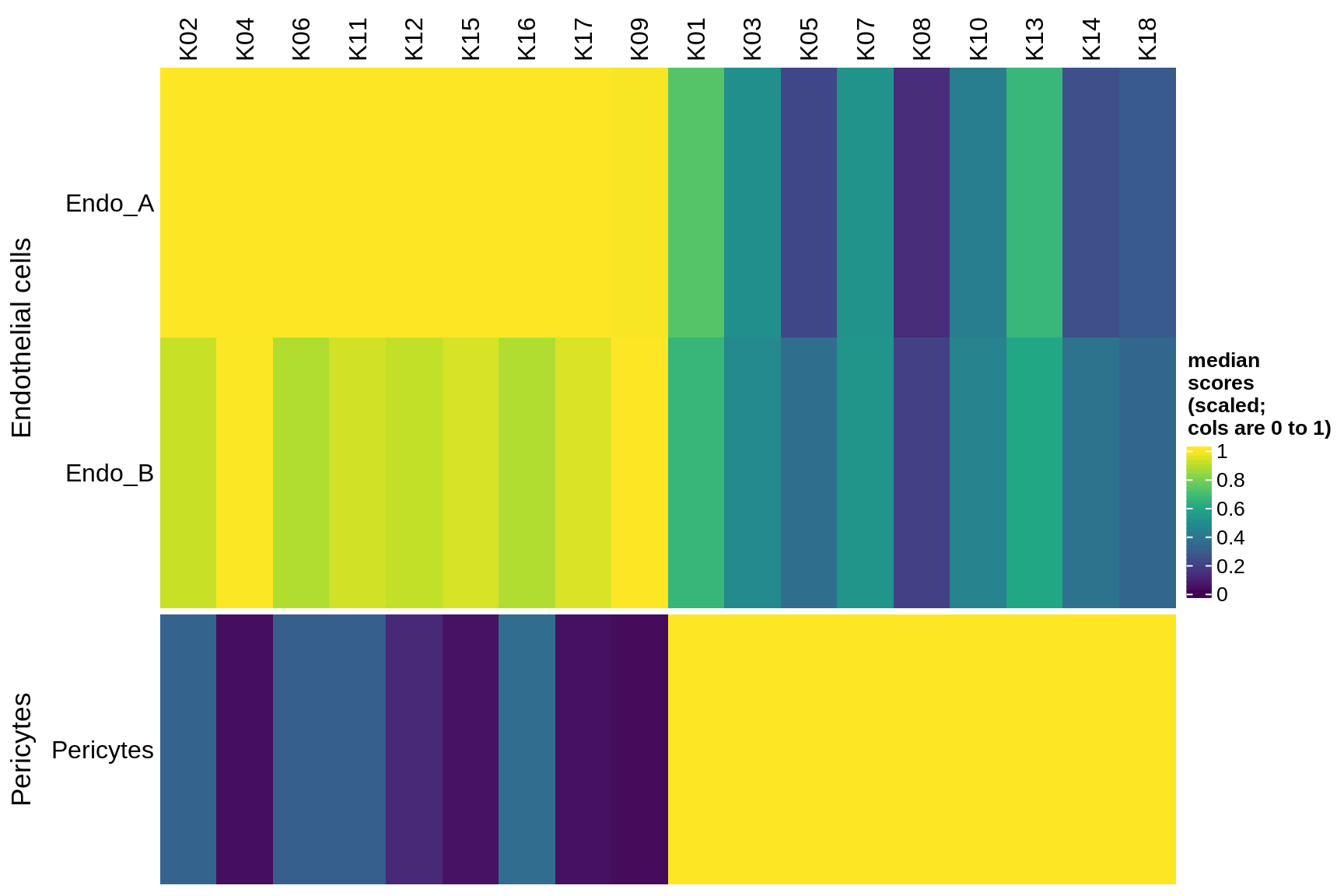
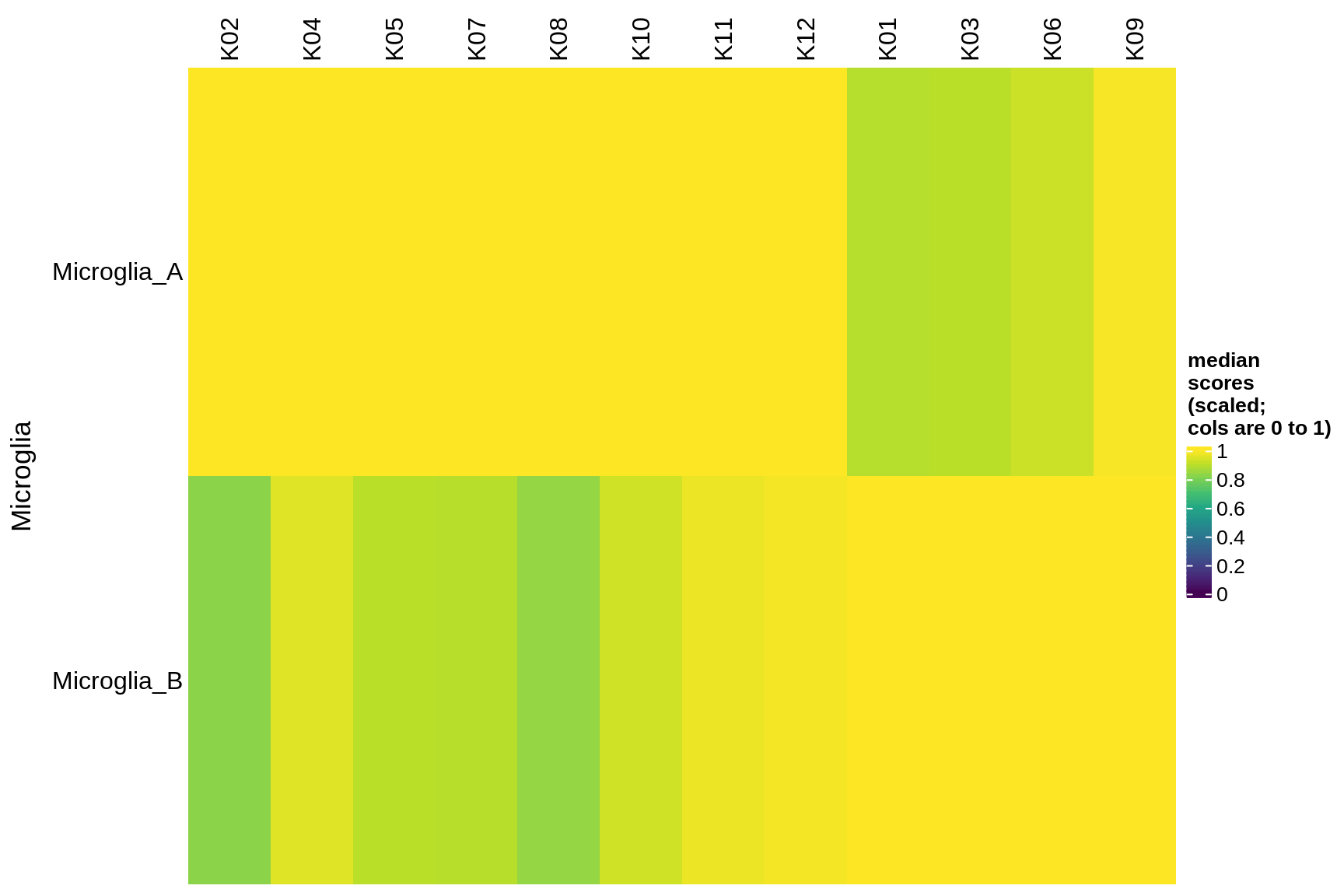
microglia
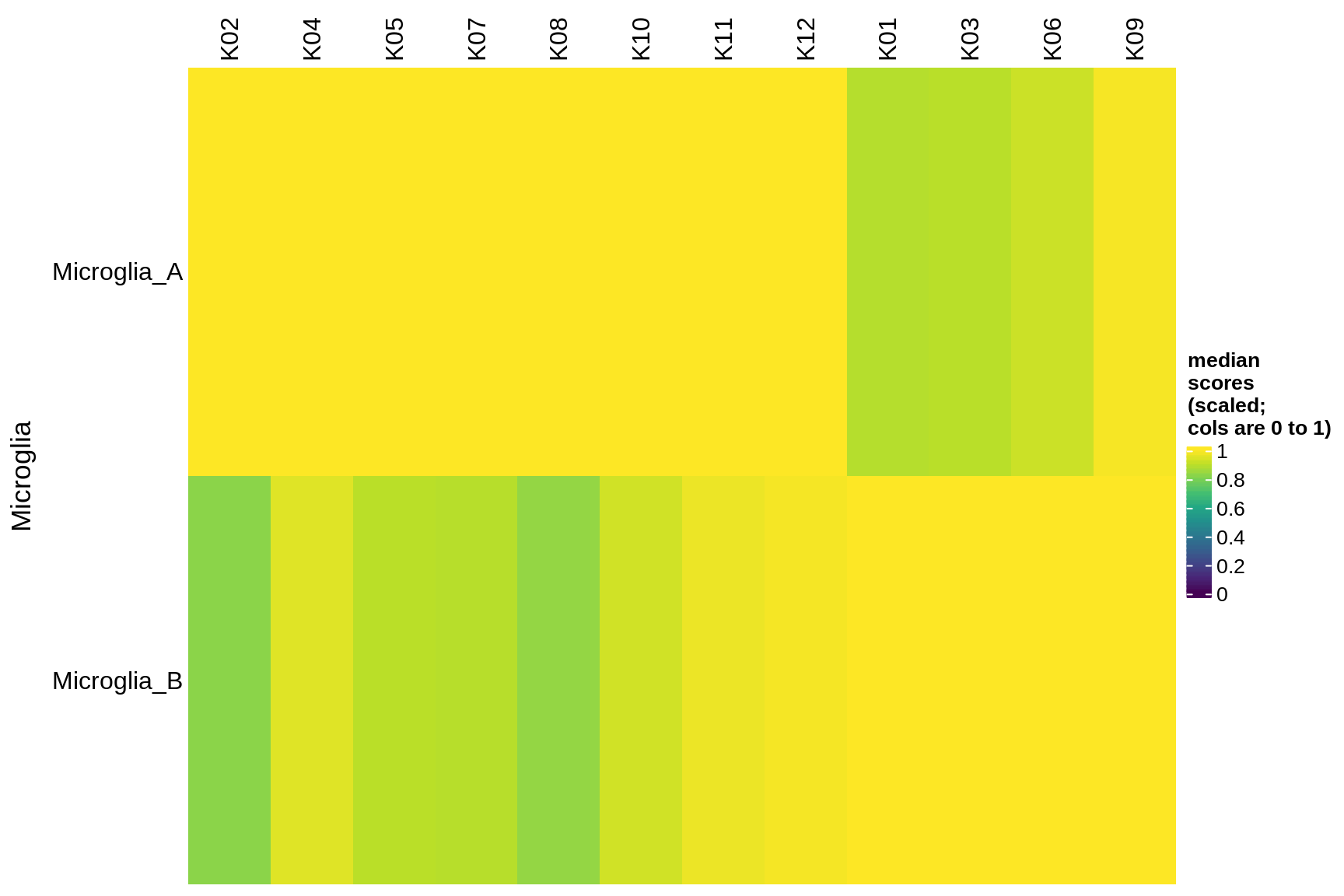
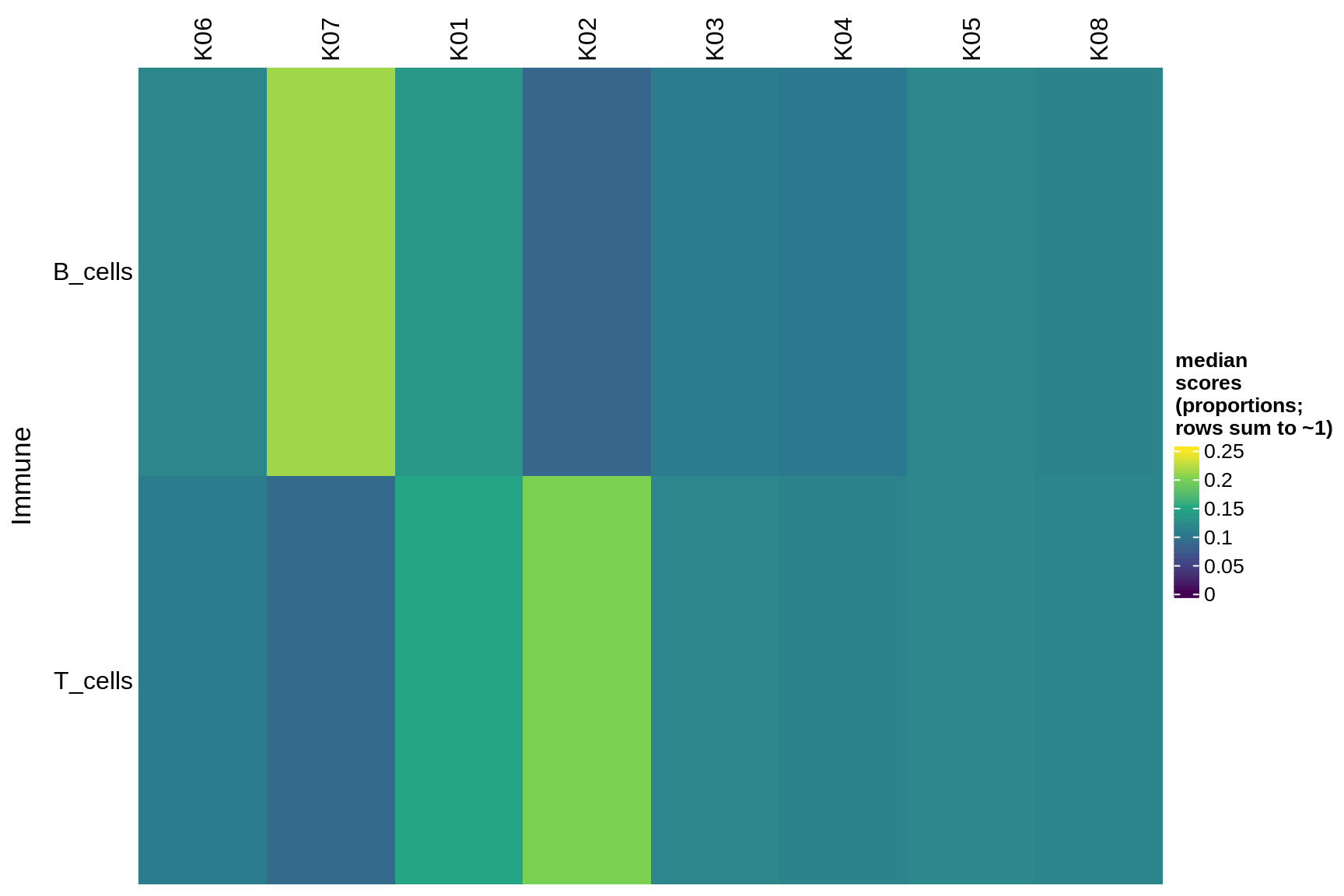
immune
Median module scores by celltype (proportions)
for (g in group_list) {
cat('### ', g, '\n')
hm = plot_scores_by_celltype(pop_list[[g]]$scores_dt, pop_list[[g]]$k_order,
what = 'propns')
if (!is.null(hm))
draw(hm)
cat('\n\n')
}oligo_opc

micro_immune
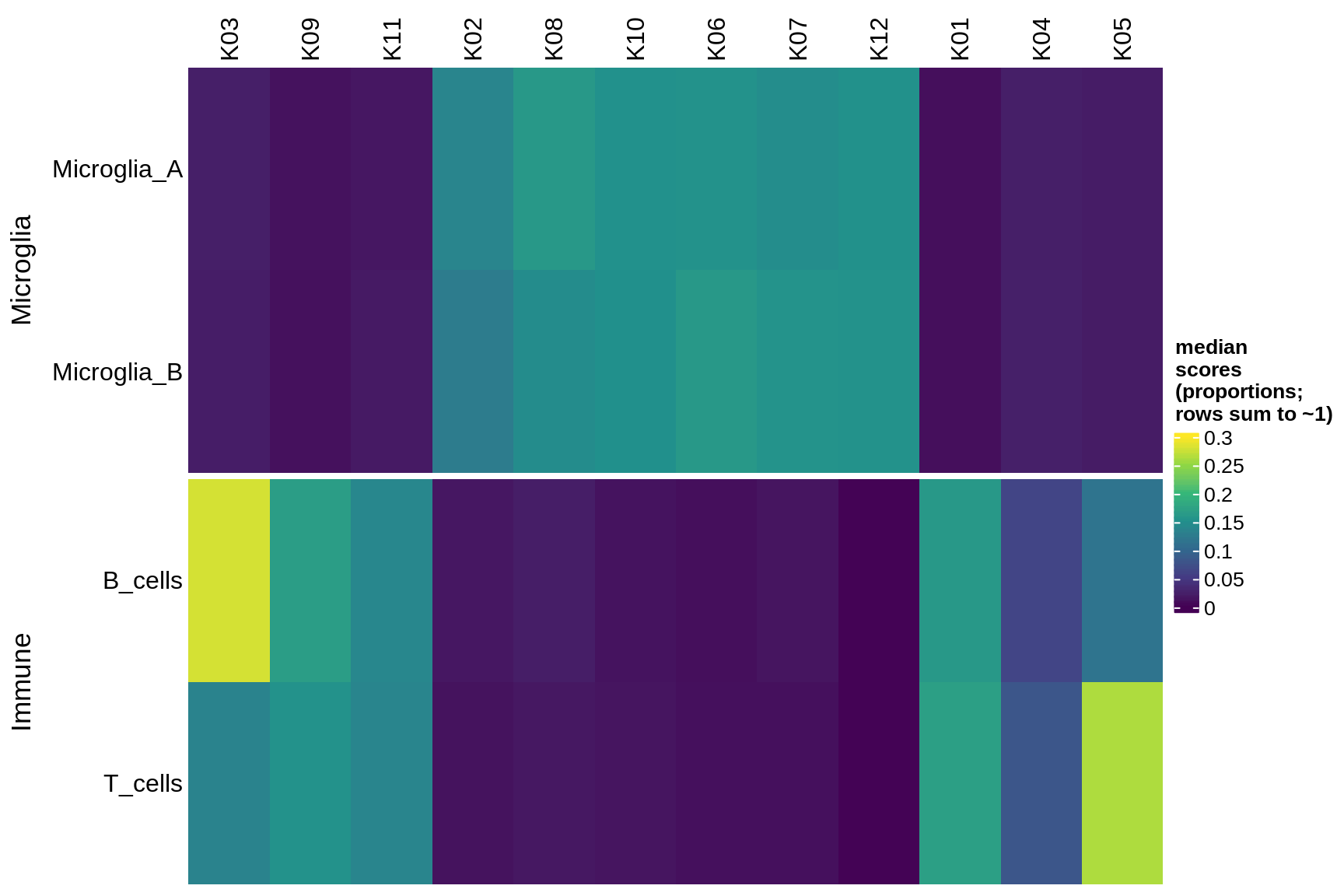
excitatory
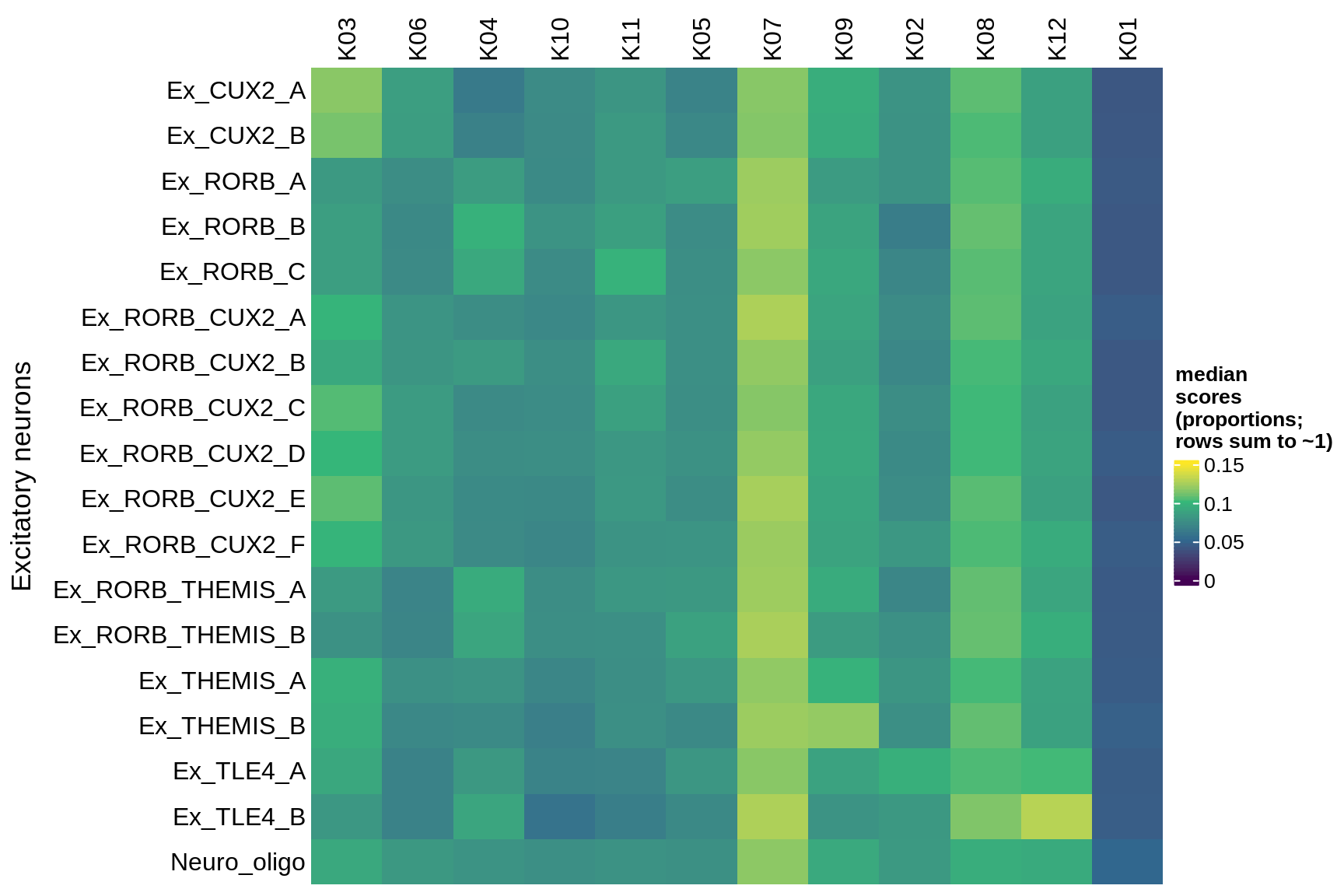
inhibitory
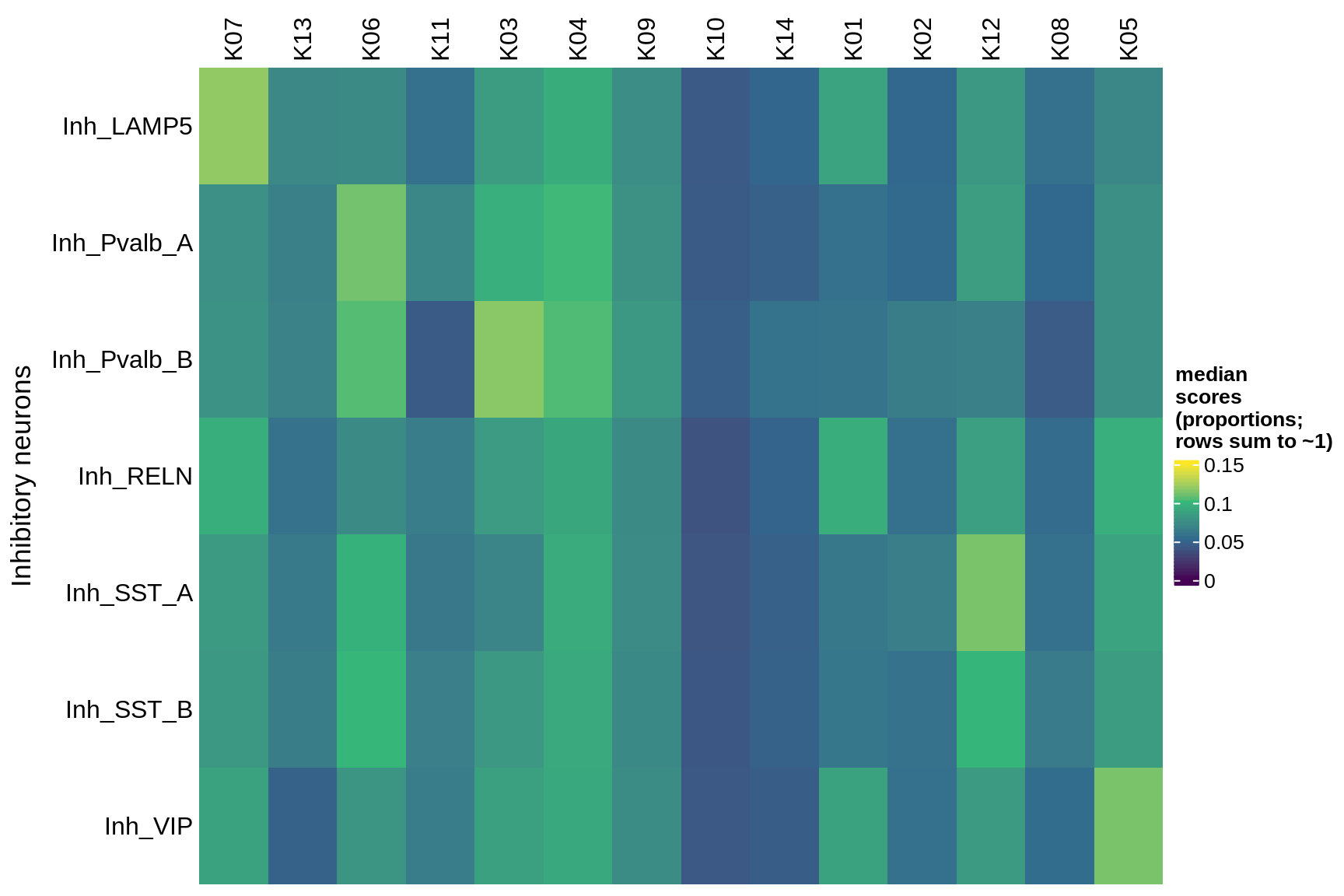
astrocytes
endo_stromal
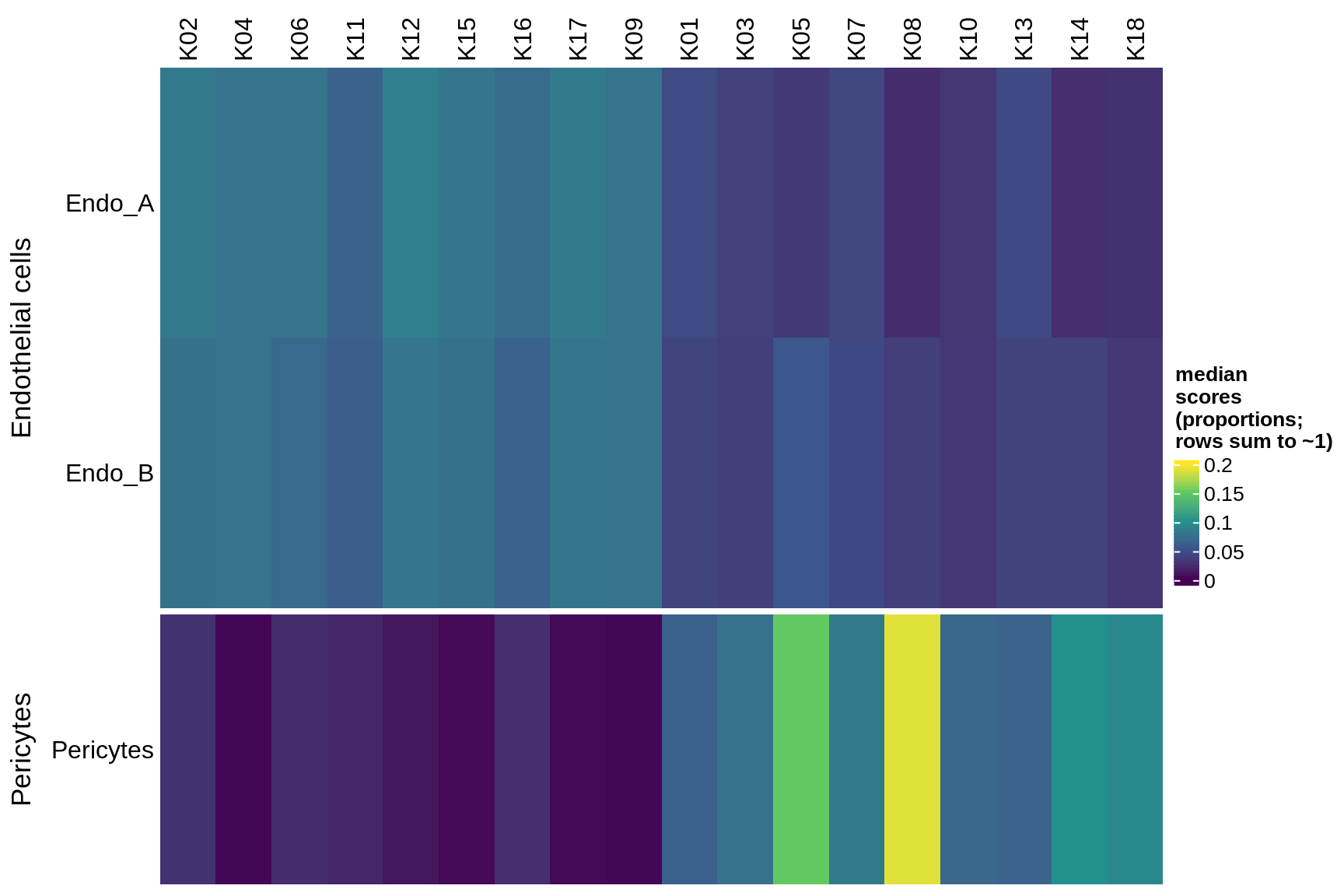
microglia
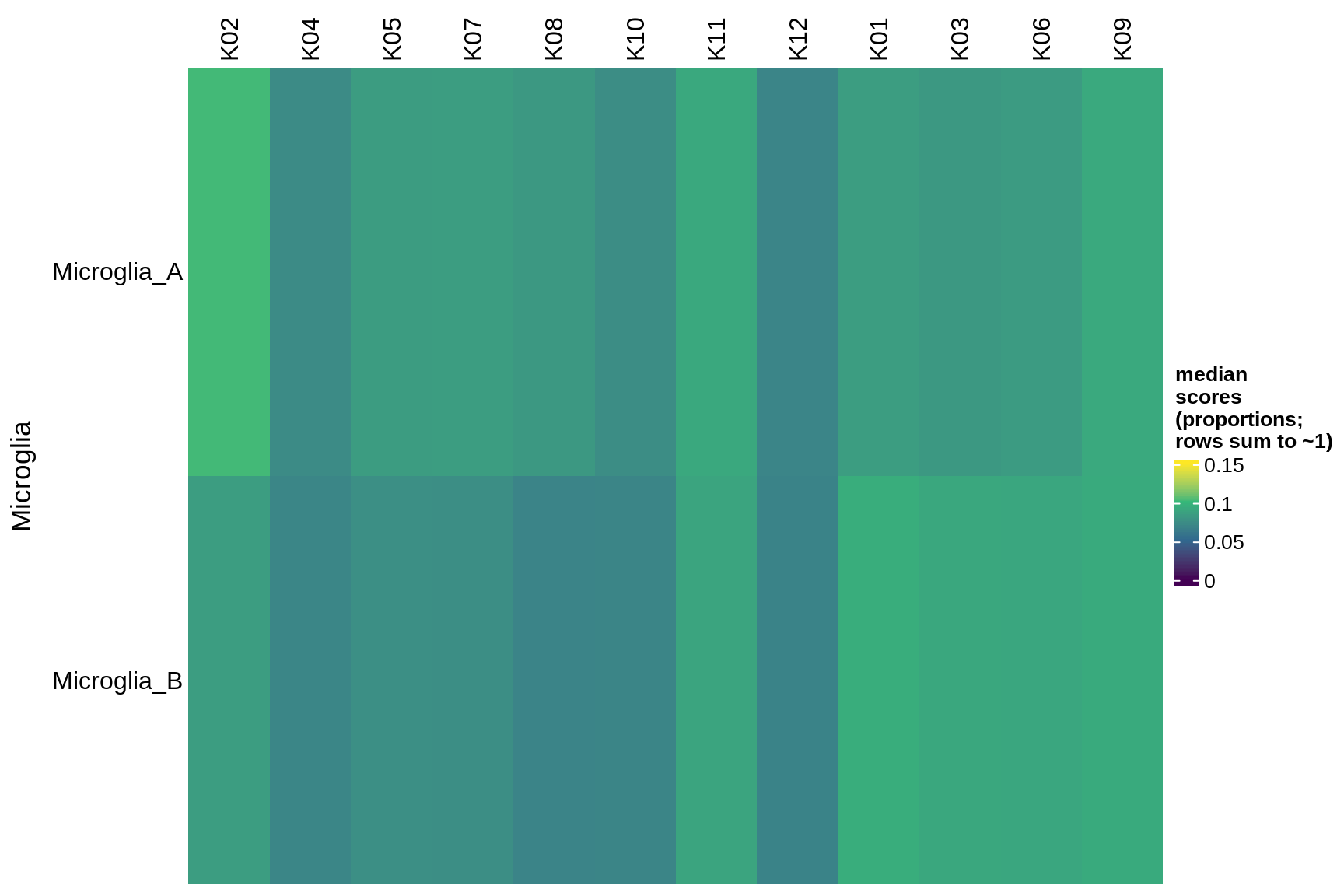
immune
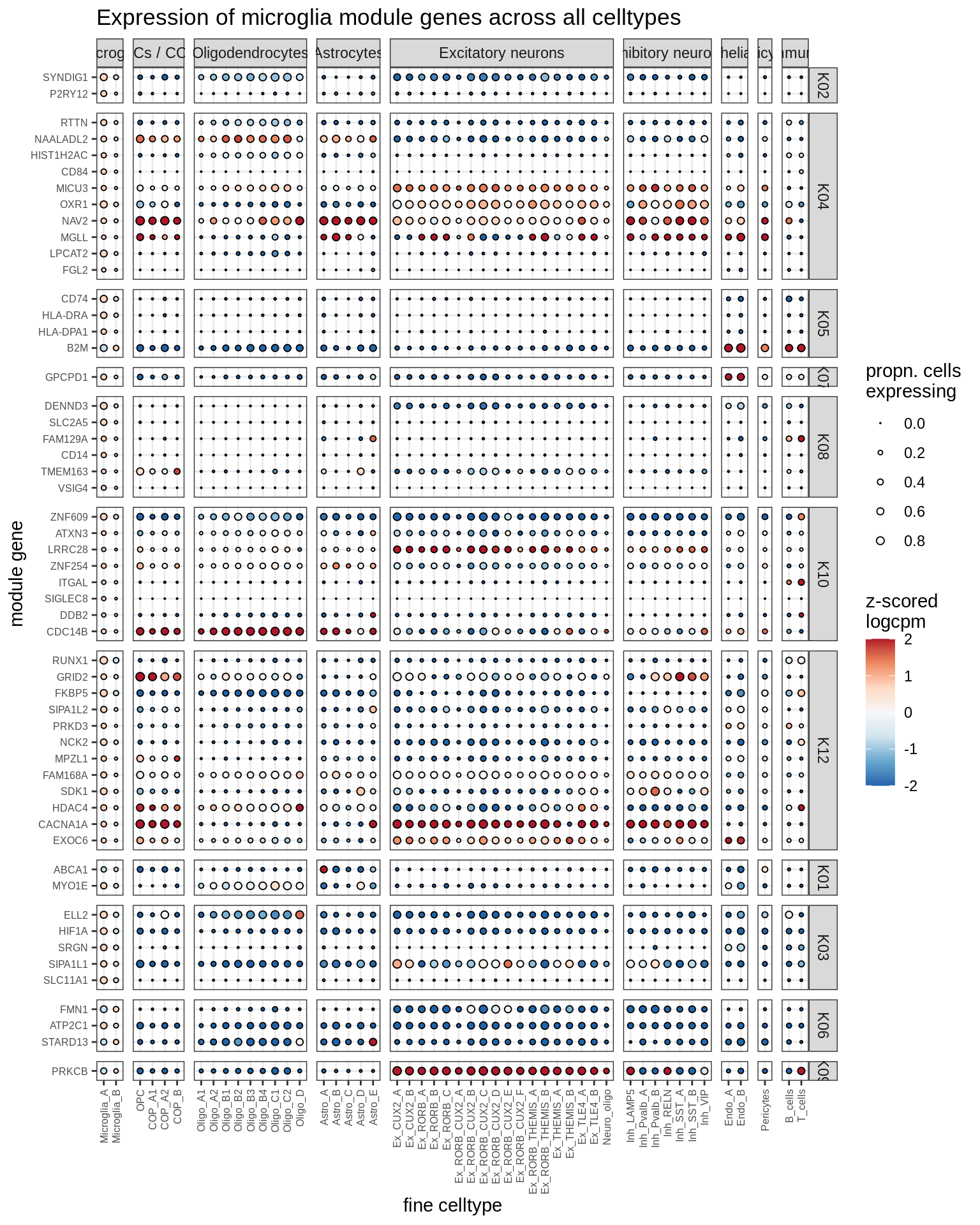
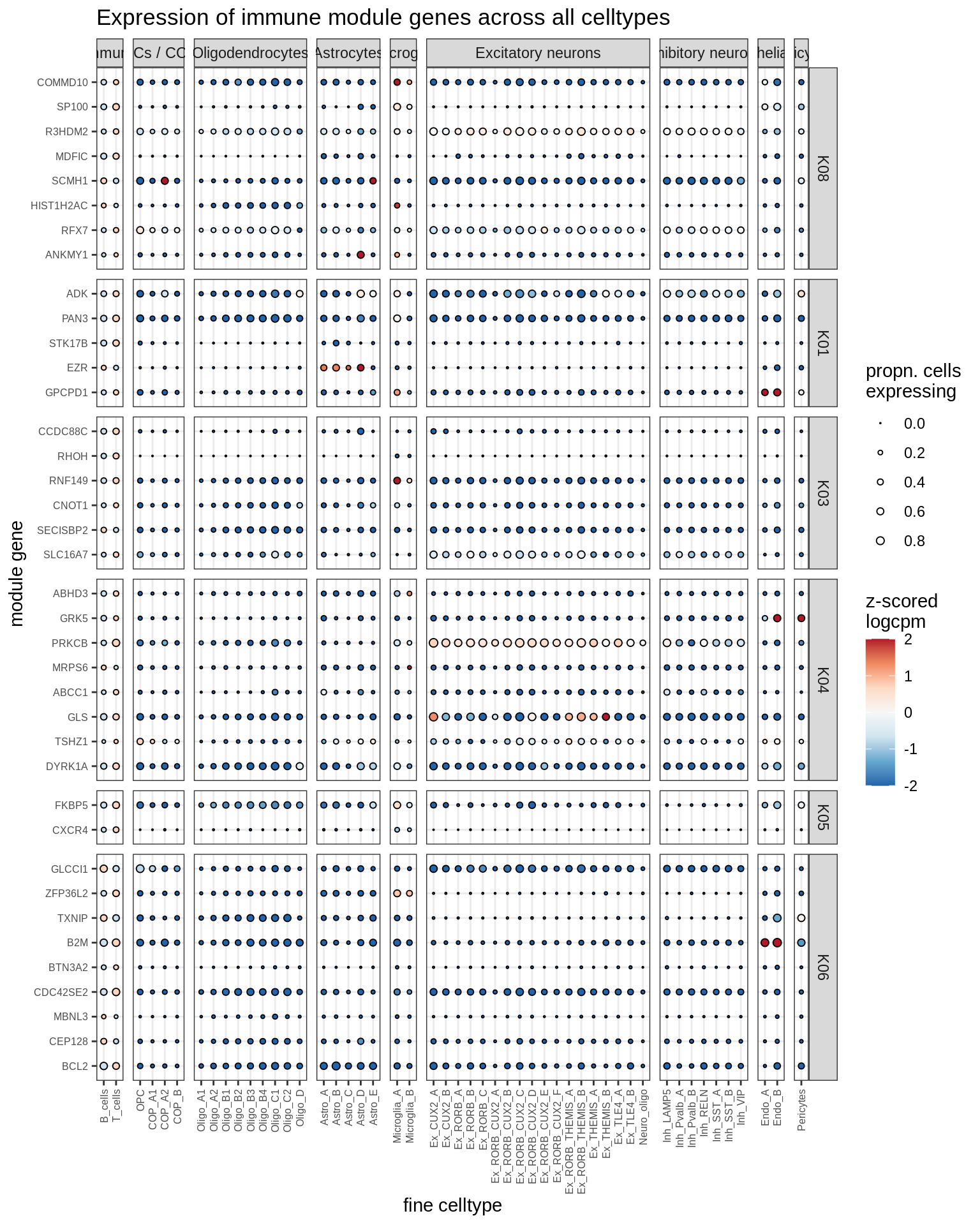
Most important genes per module, dotplot
for (g in group_list) {
cat('### ', g, '\n')
print(plot_biggest_genes_dotplot(g, spec_list, pop_list[[g]]$w_mat,
pop_list[[g]]$k_order, fine_dt, w2_cut = 0.02))
cat('\n\n')
}oligo_opc
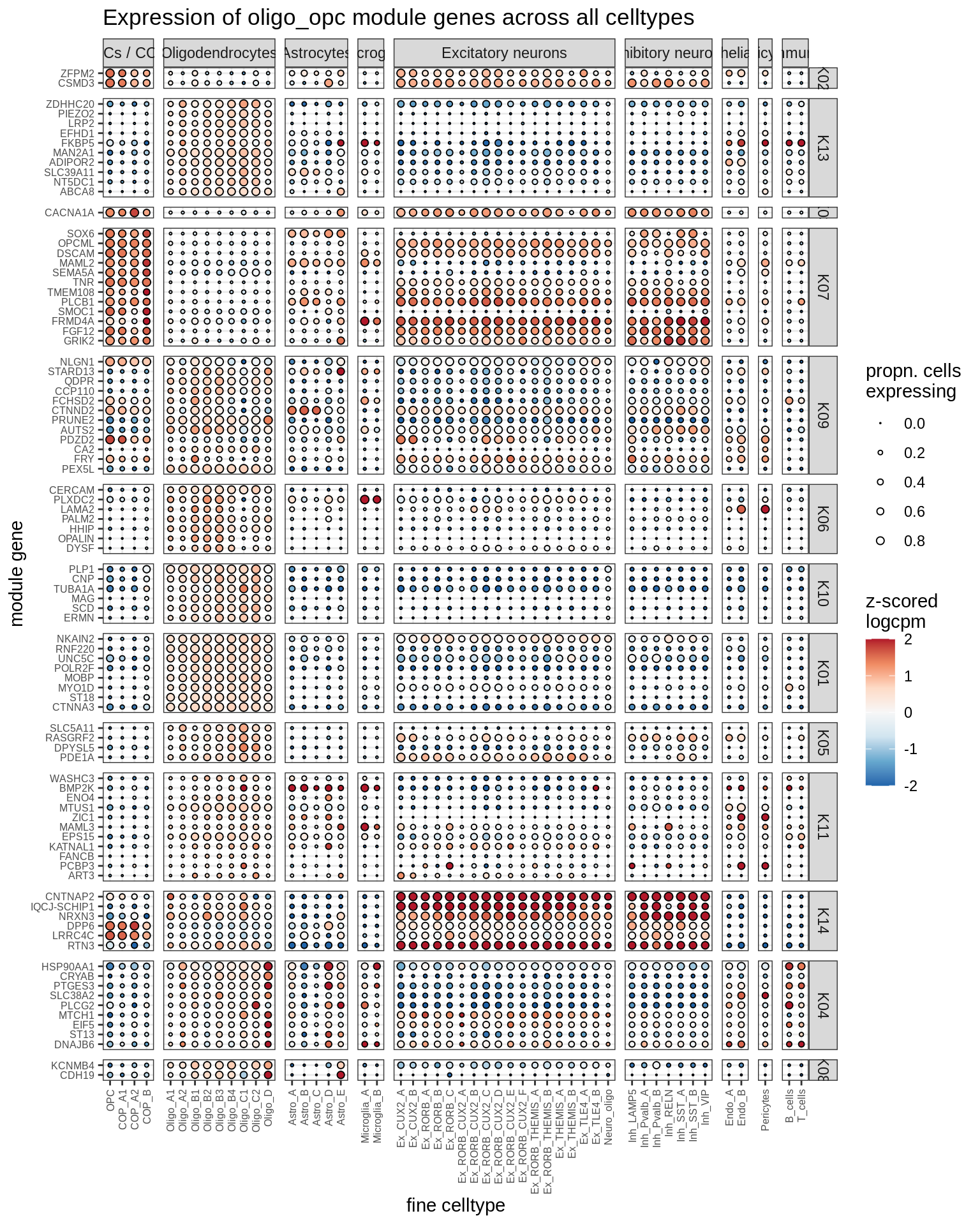
micro_immune
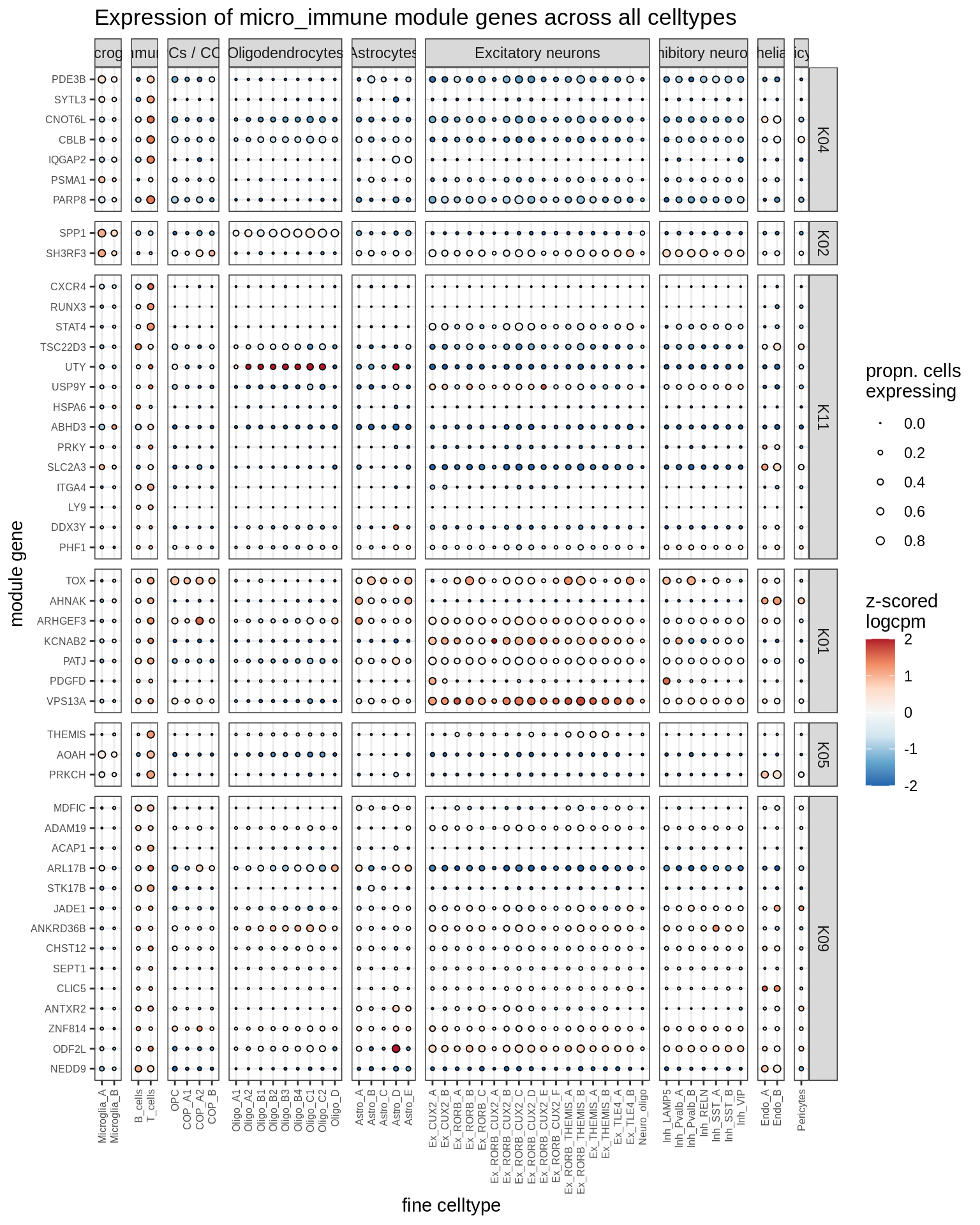
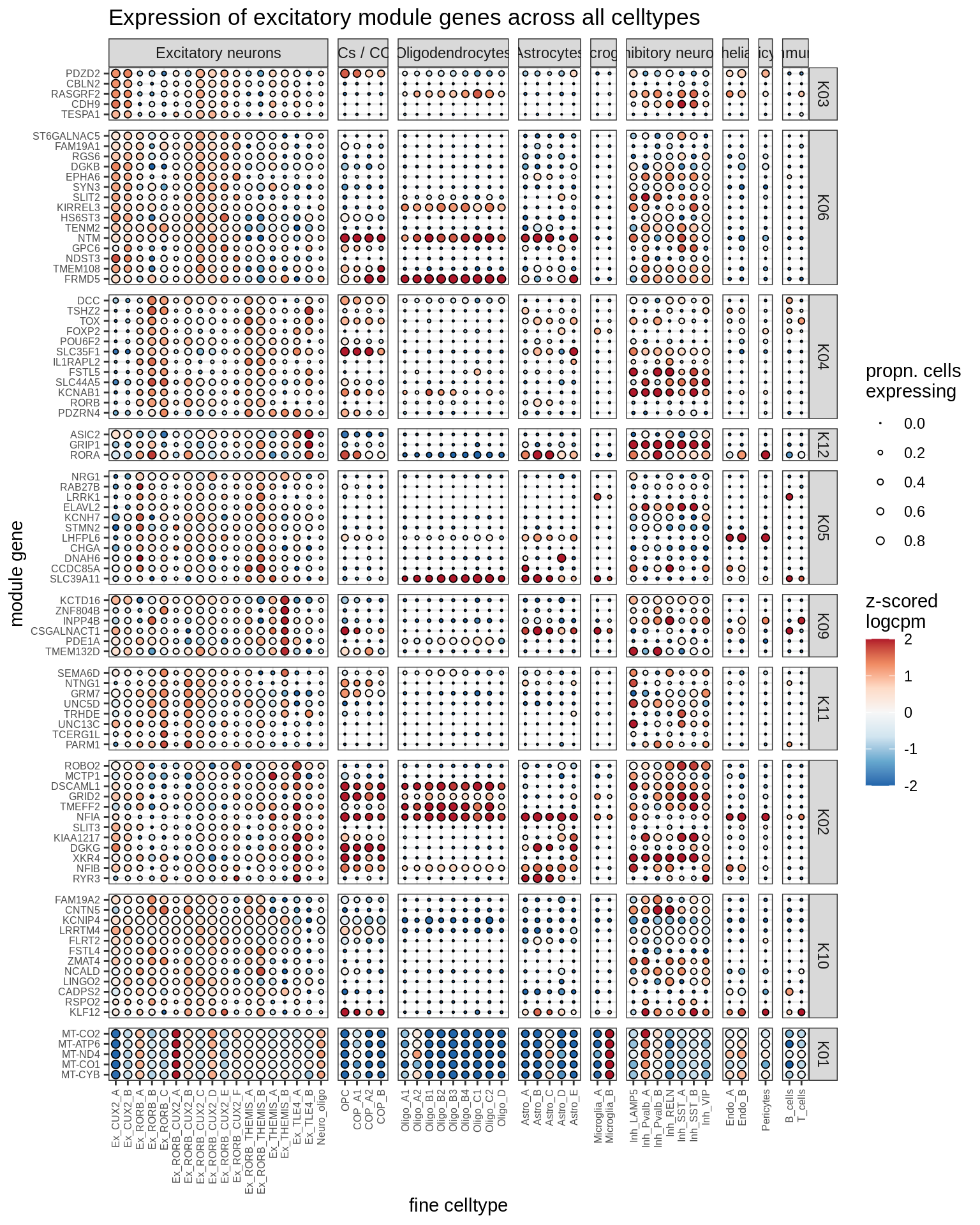
excitatory
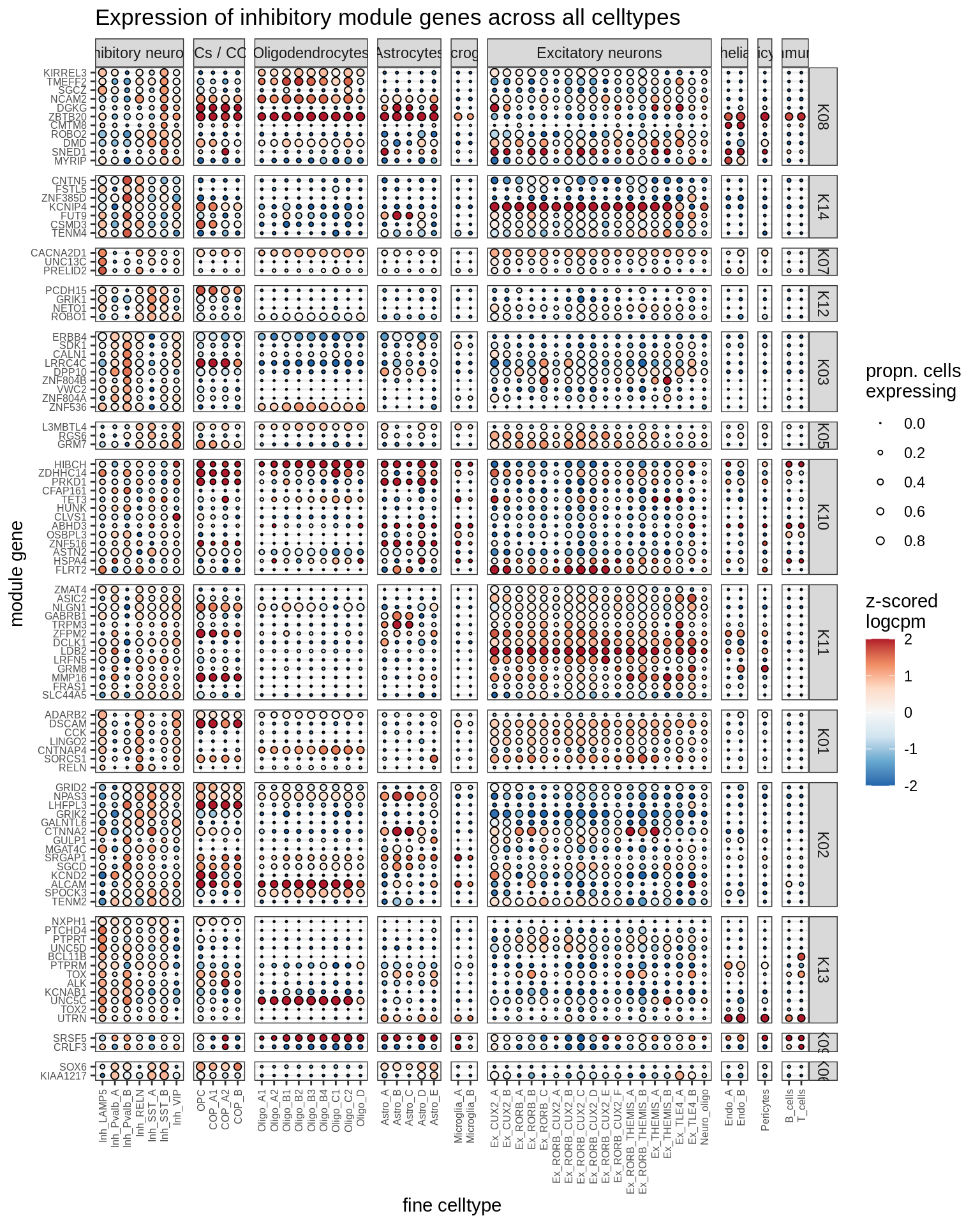
inhibitory
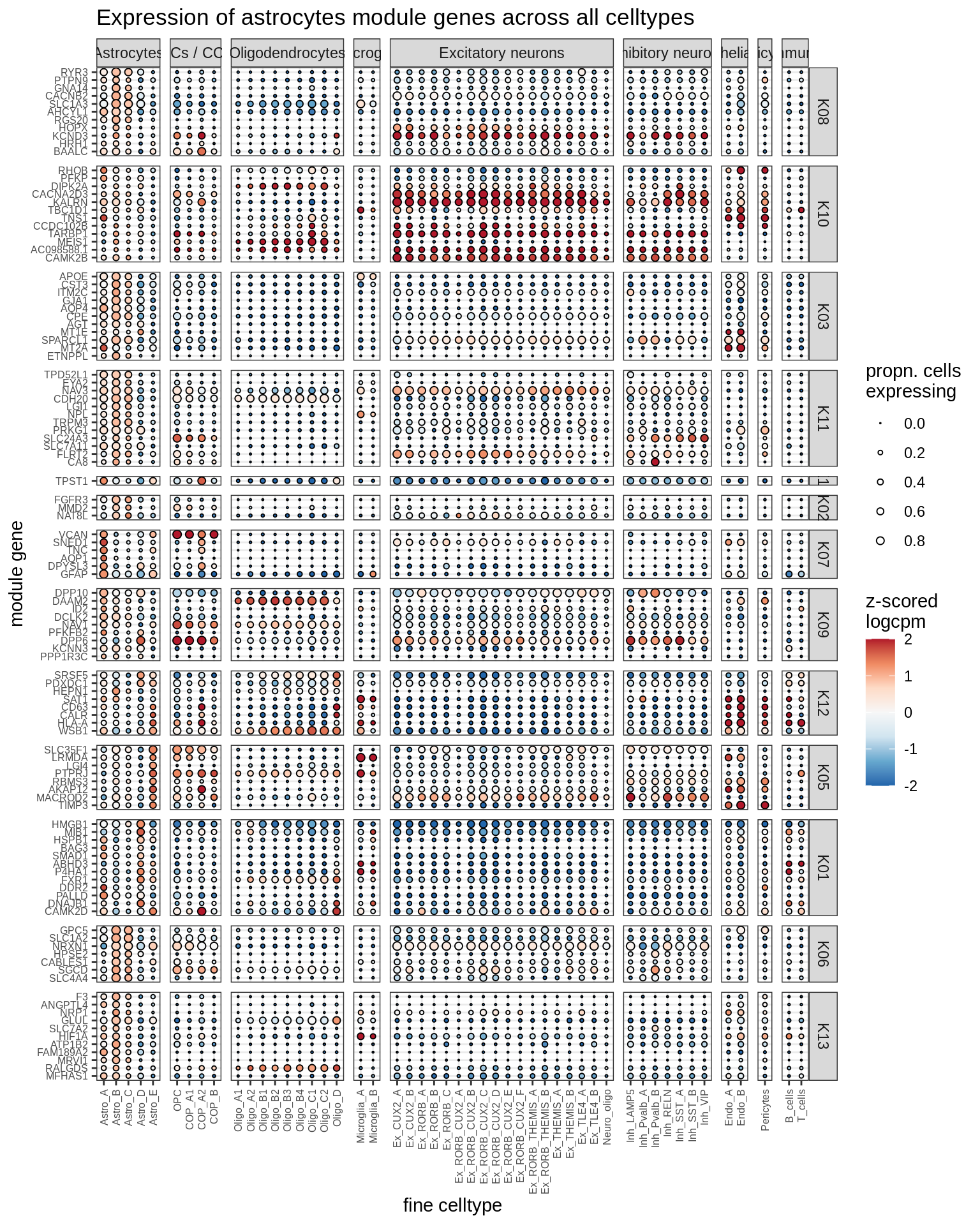
astrocytes
endo_stromal
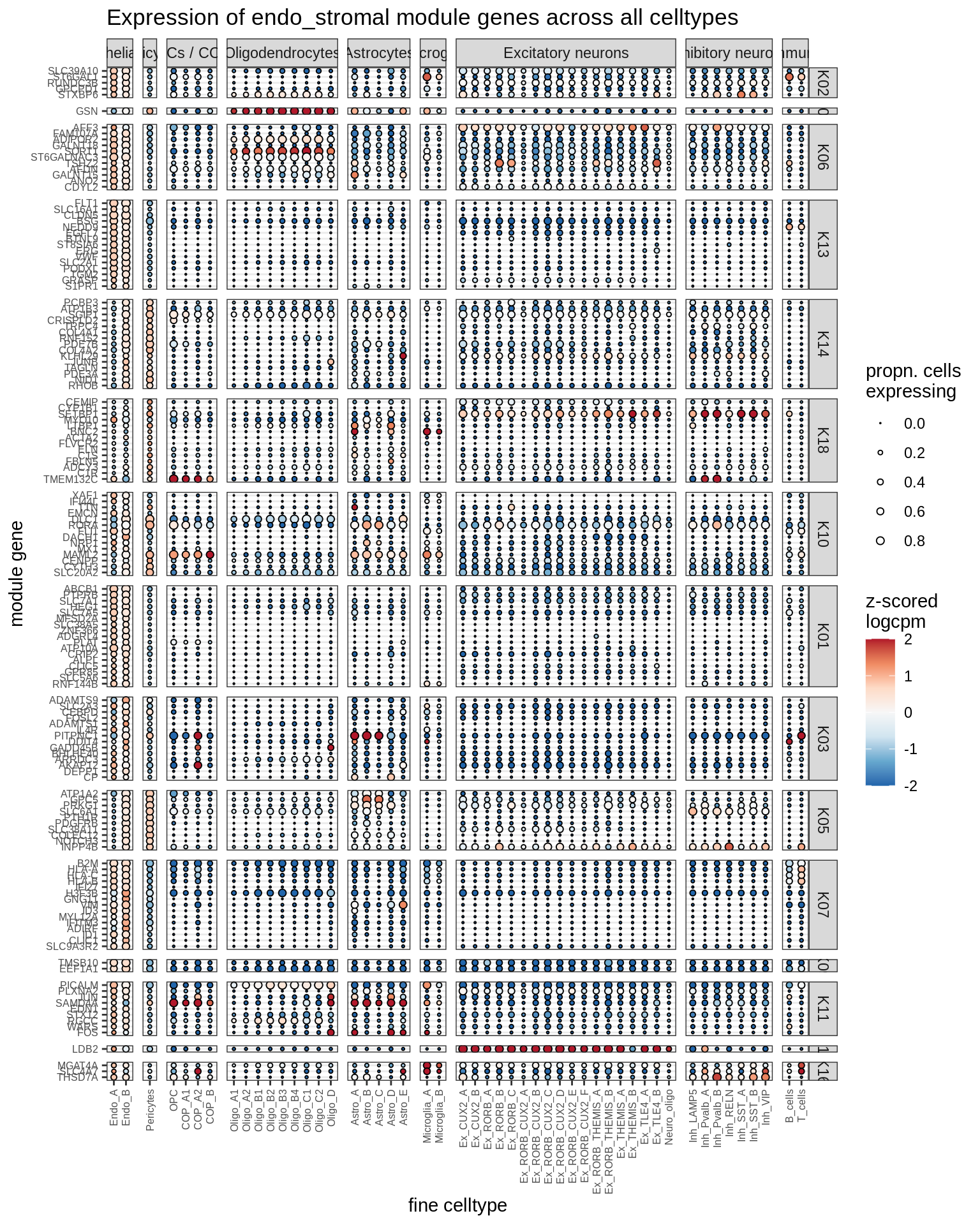
microglia
immune
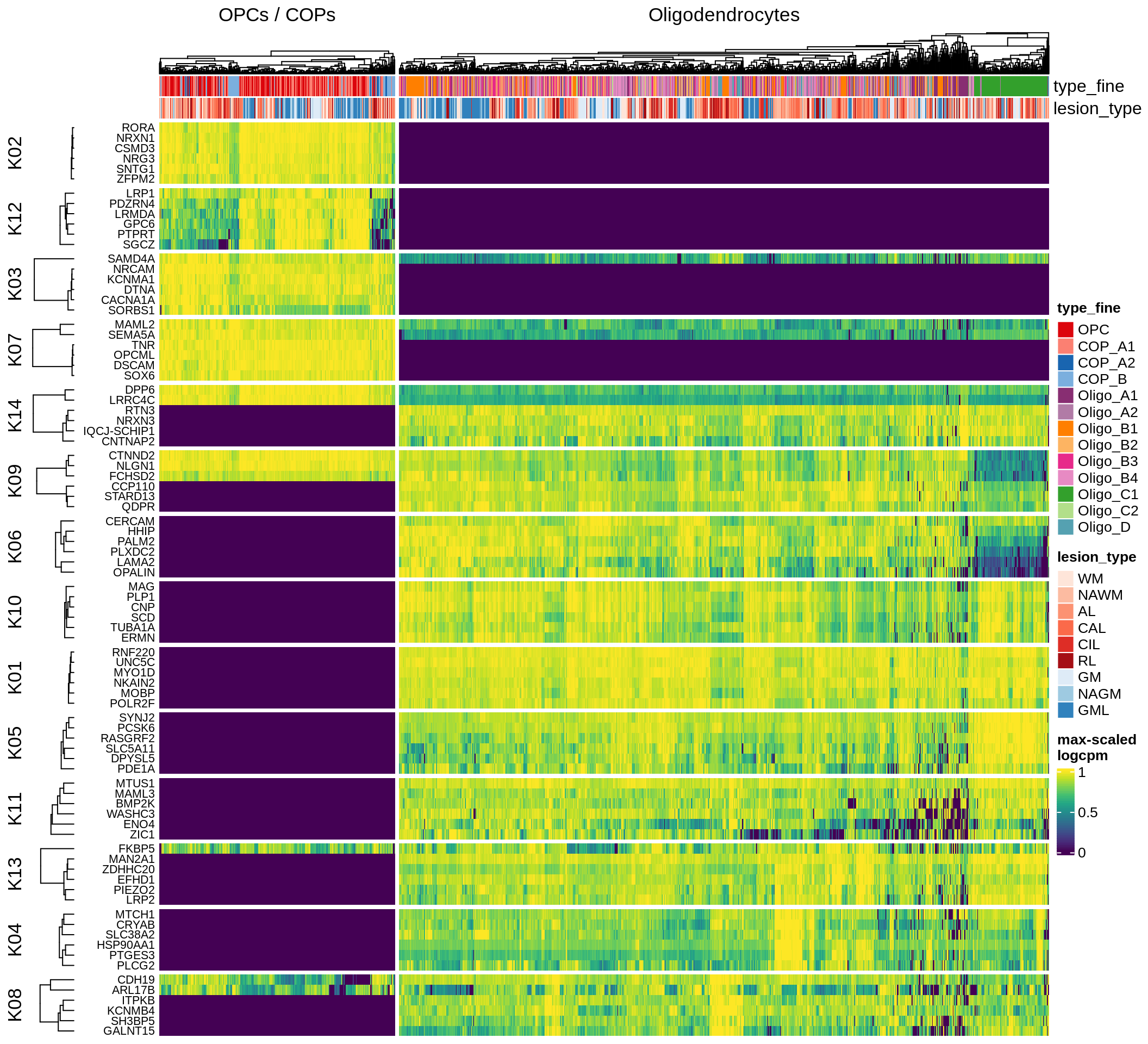
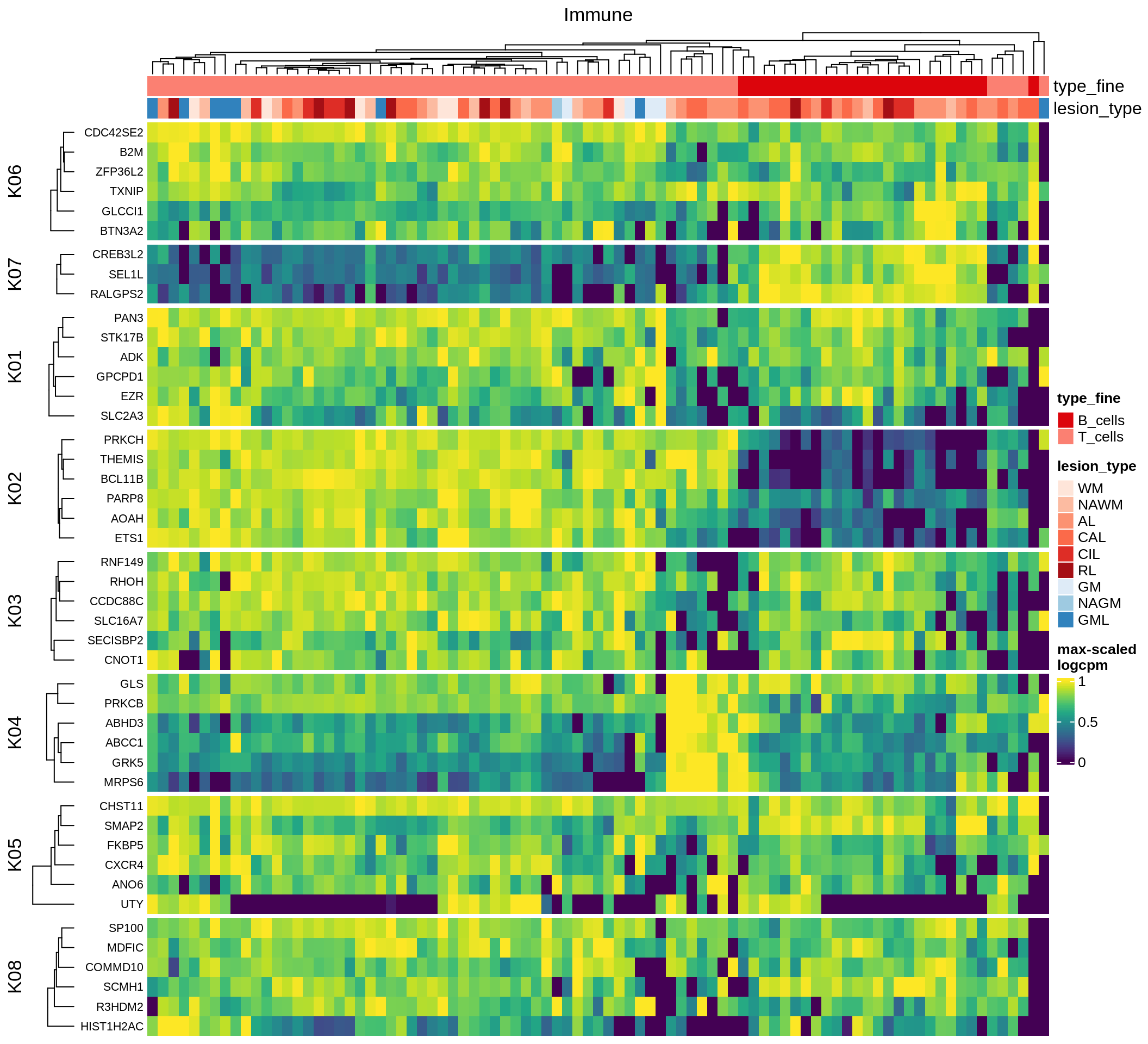
Most important genes per module, heatmap
for (g in group_list) {
cat('### ', g, '\n')
hm = plot_biggest_genes_heatmap(g, sce_pat, pop_list[[g]]$w_mat,
pop_list[[g]]$k_order, meta_dt, labels_dt, w2_cut = 0.01)
if (!is.null(hm))
draw(hm, merge_legend = TRUE)
cat('\n\n')
}oligo_opc
micro_immune
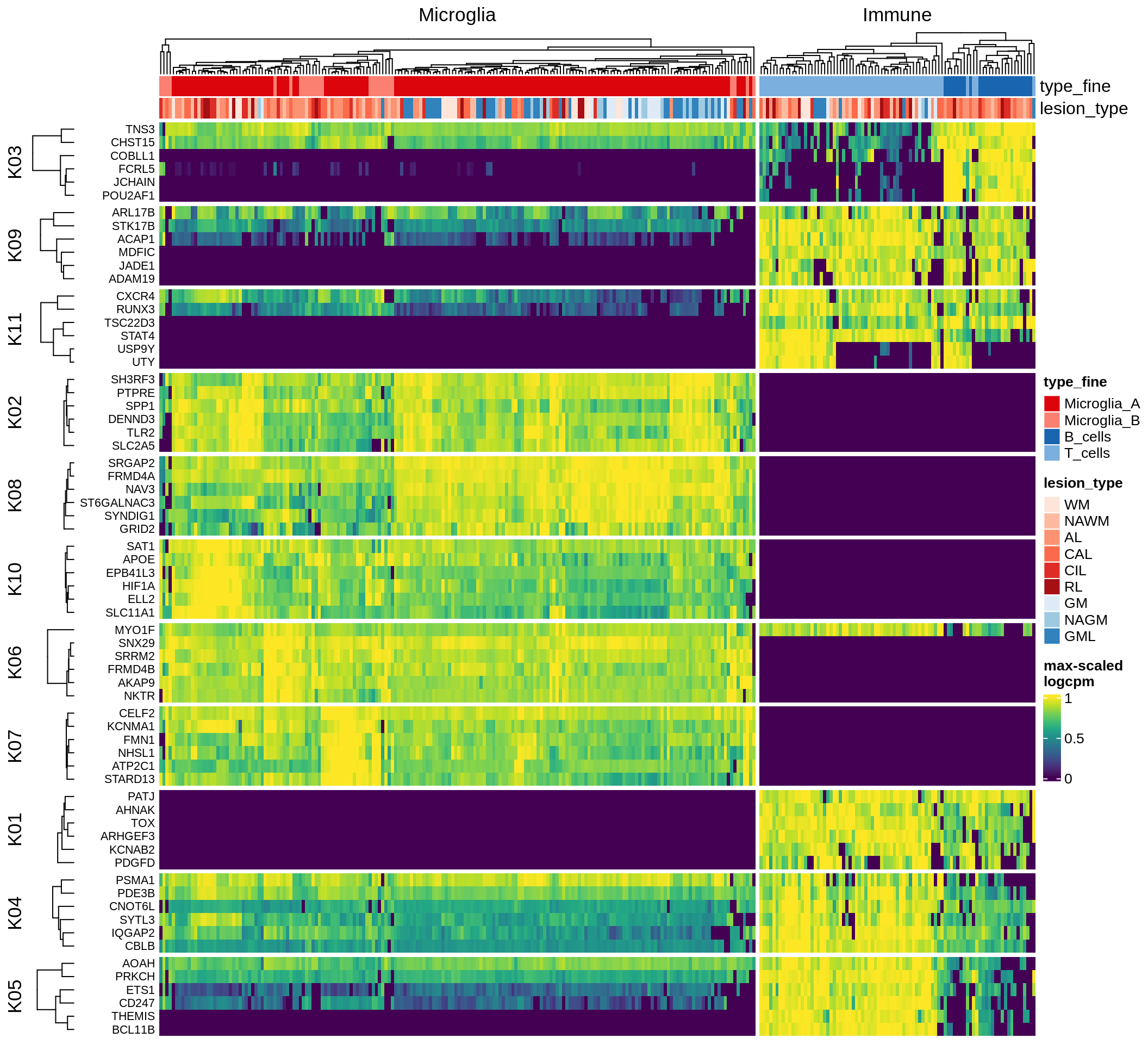
excitatory
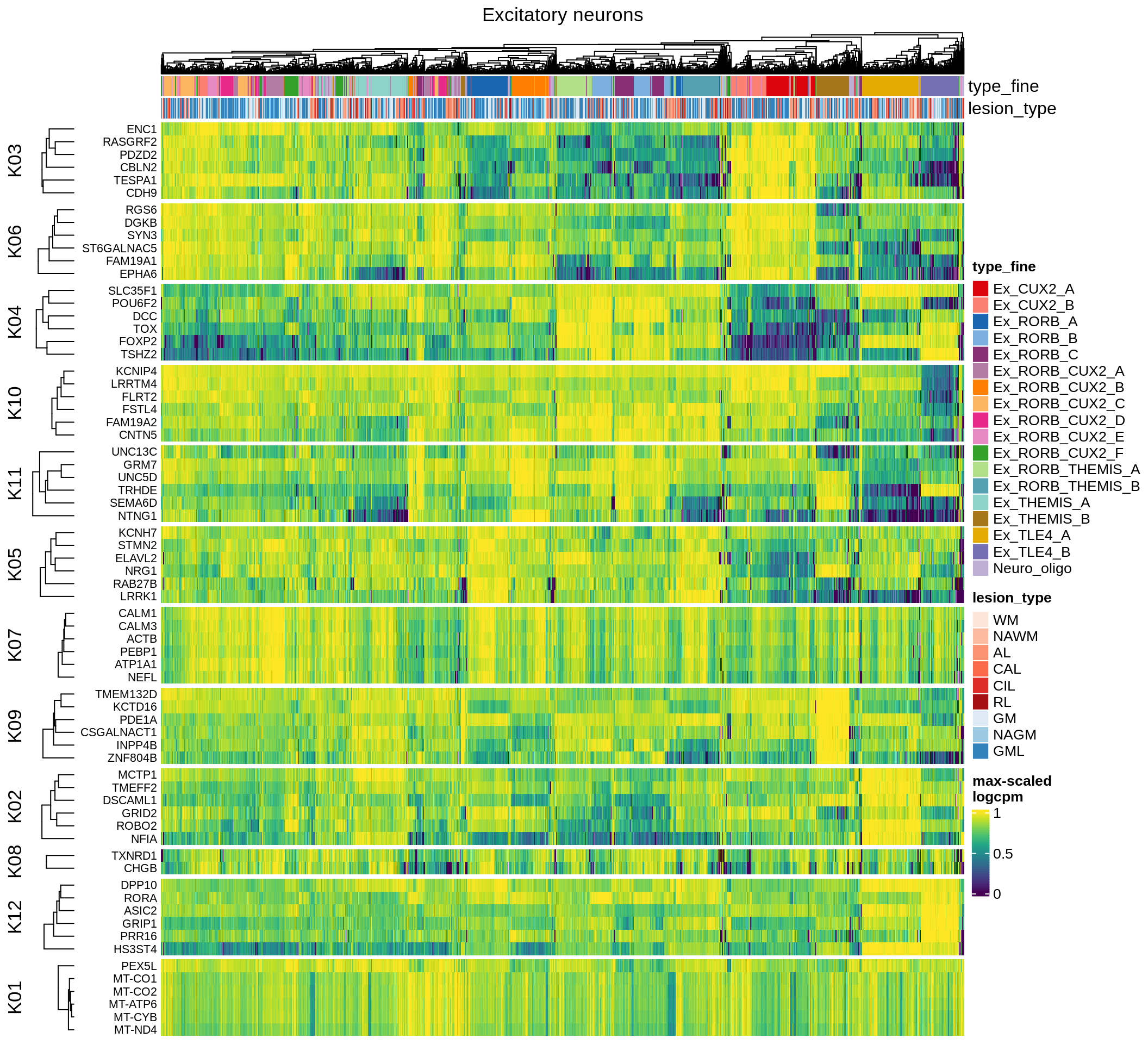
inhibitory
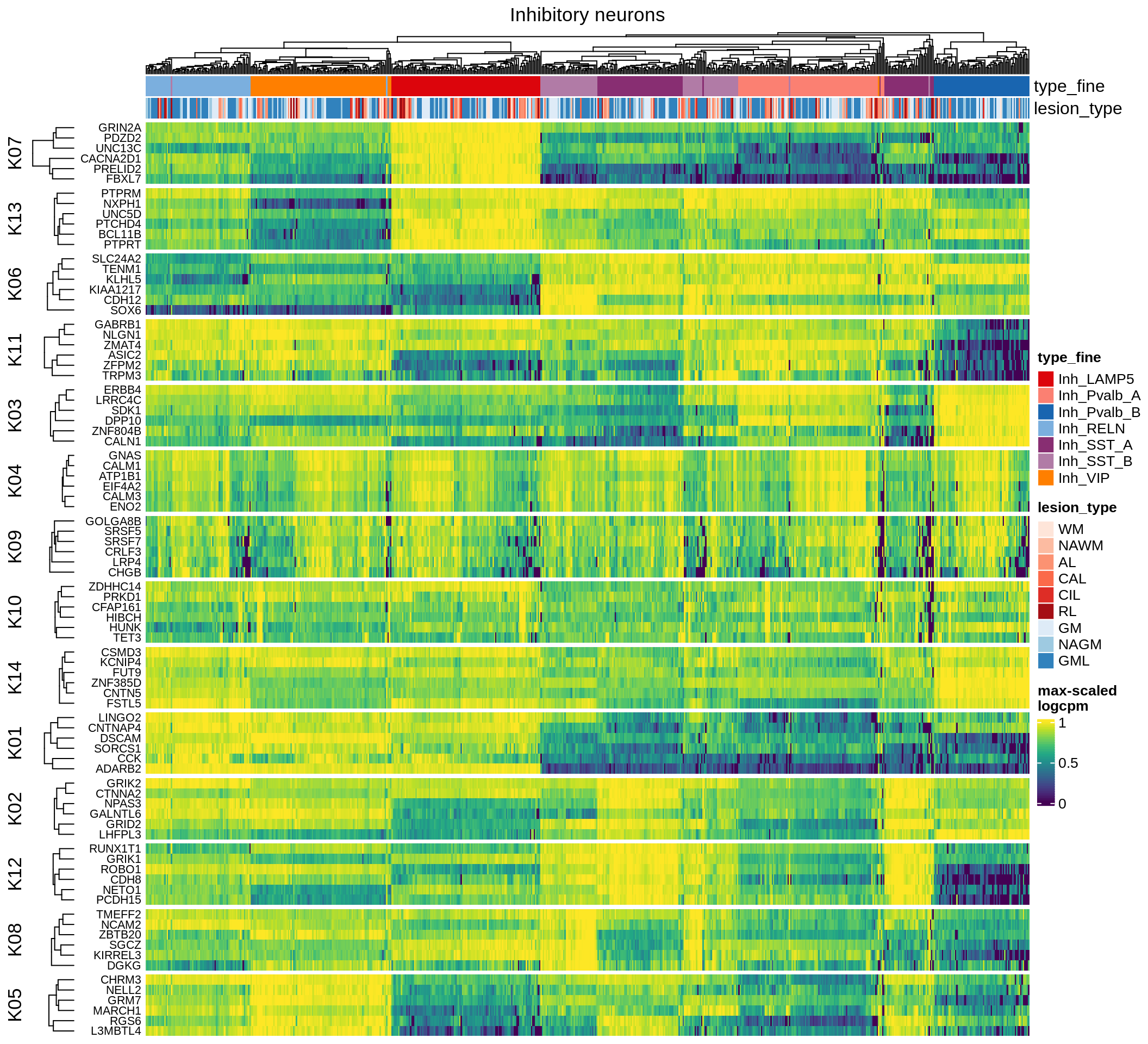
astrocytes
endo_stromal
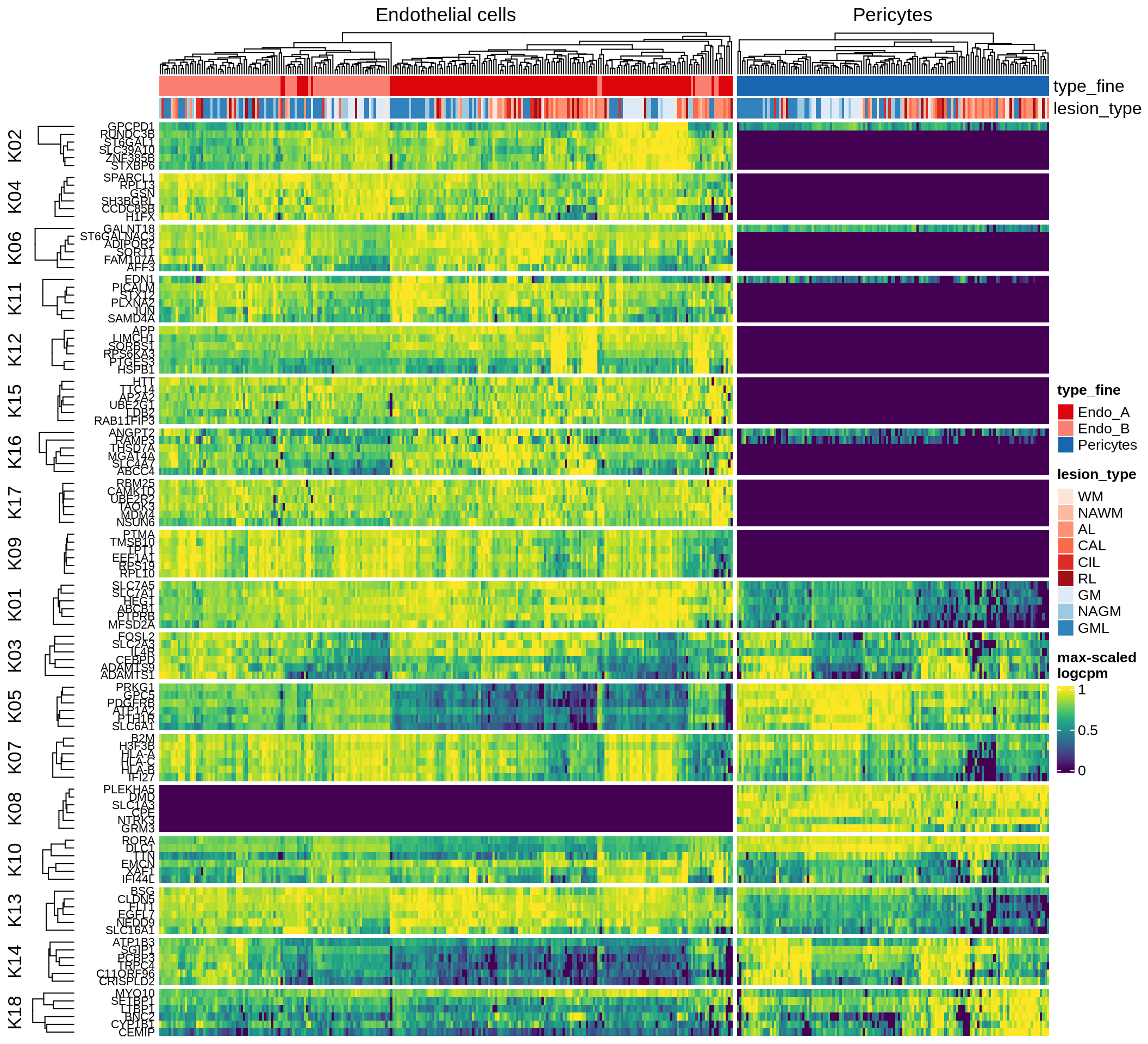
microglia
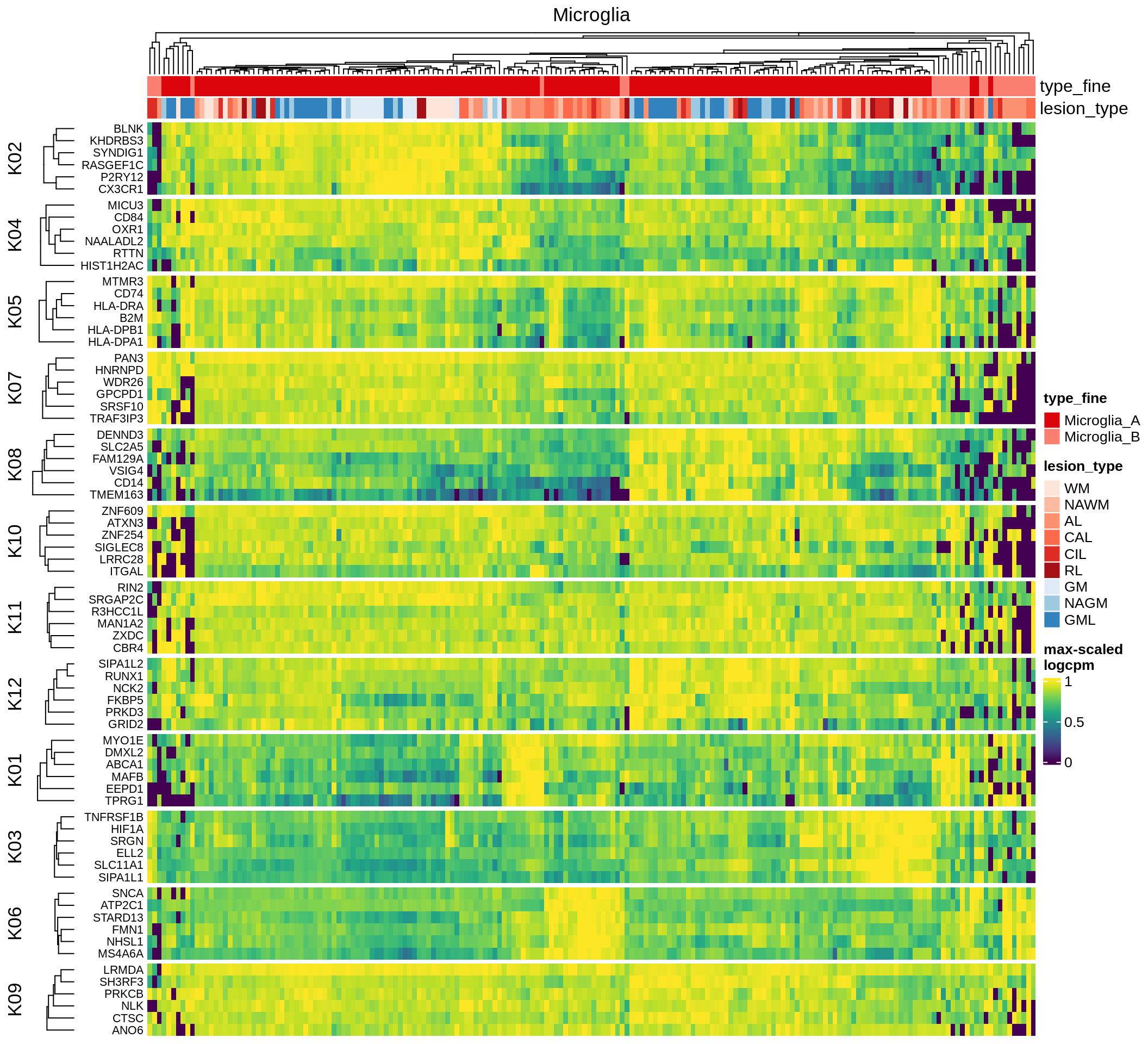
immune
Median module scores by celltype (scaled)
for (g in group_list) {
cat('### ', g, '\n')
hm = plot_scores_by_celltype(pop_list[[g]]$scores_dt, pop_list[[g]]$k_order,
what = 'scaled')
if (!is.null(hm))
draw(hm)
cat('\n\n')
}oligo_opc
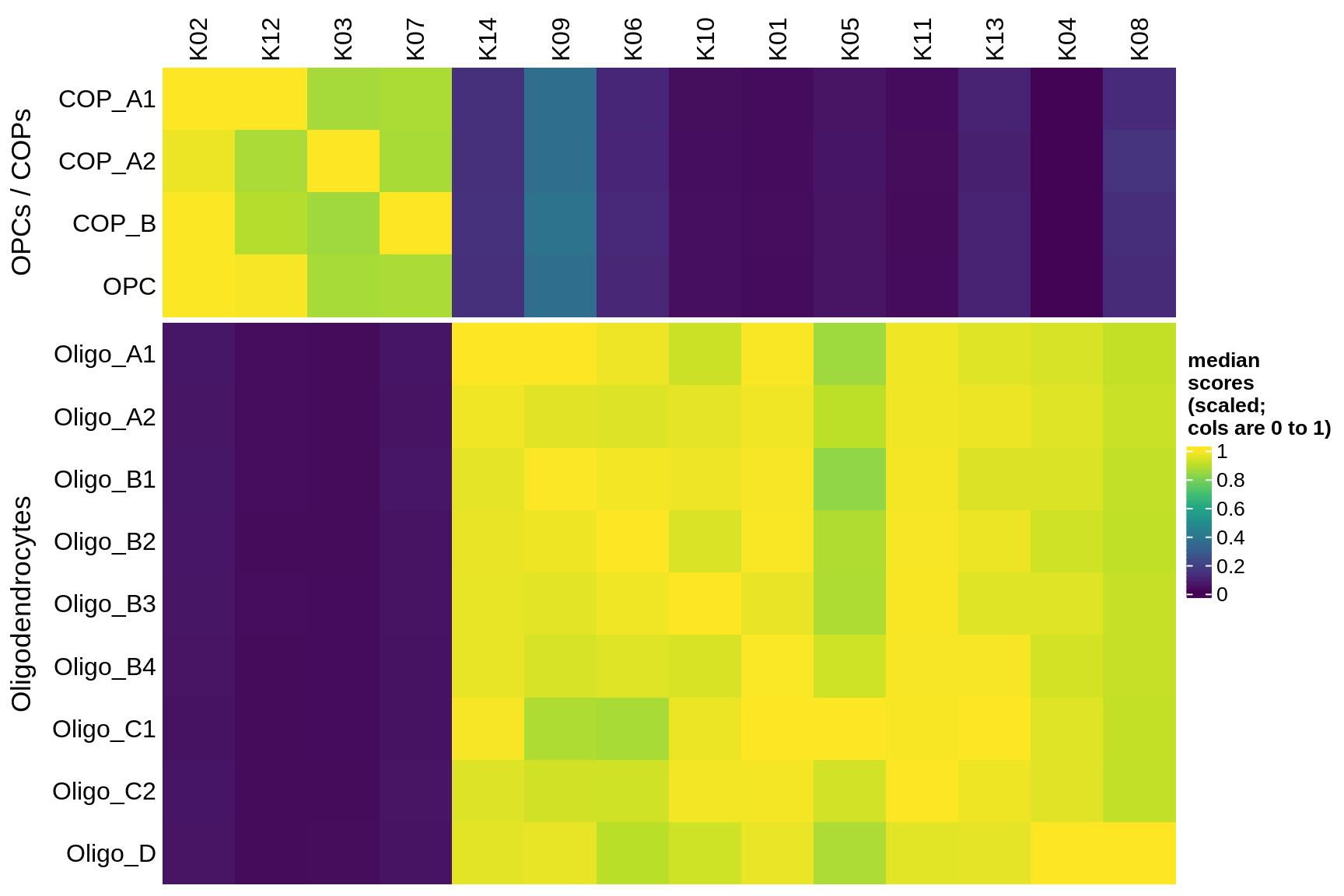
micro_immune
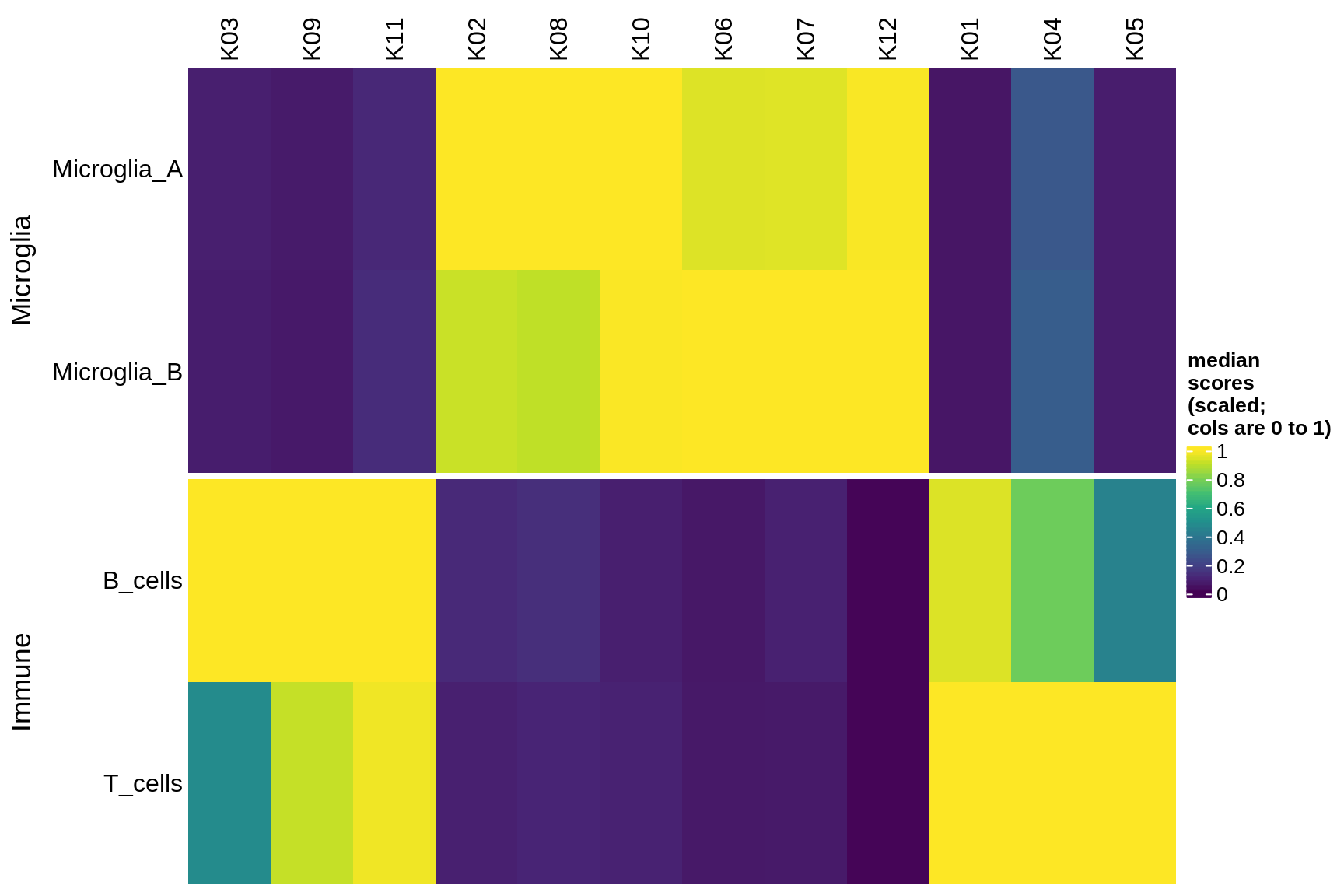
excitatory
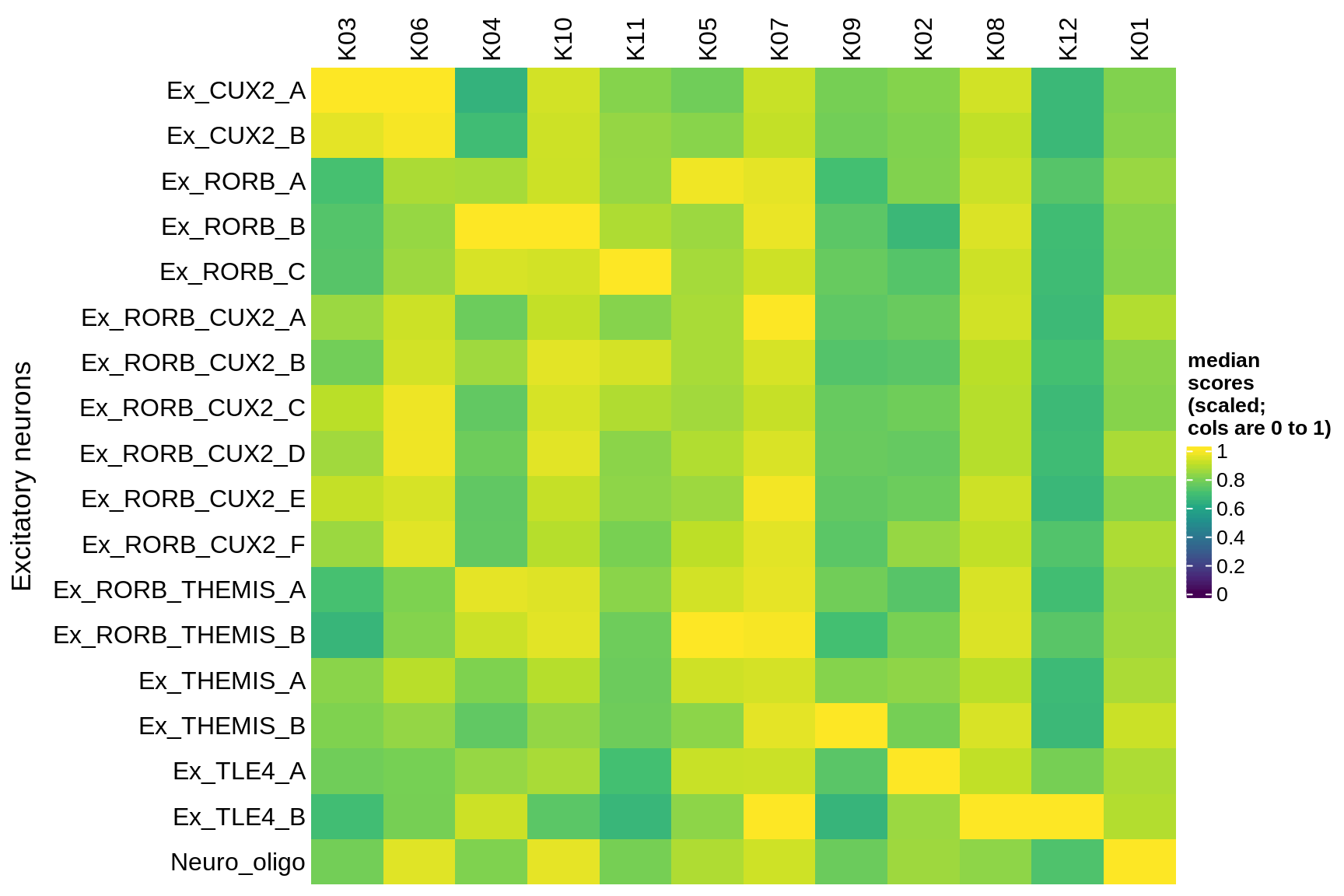
inhibitory
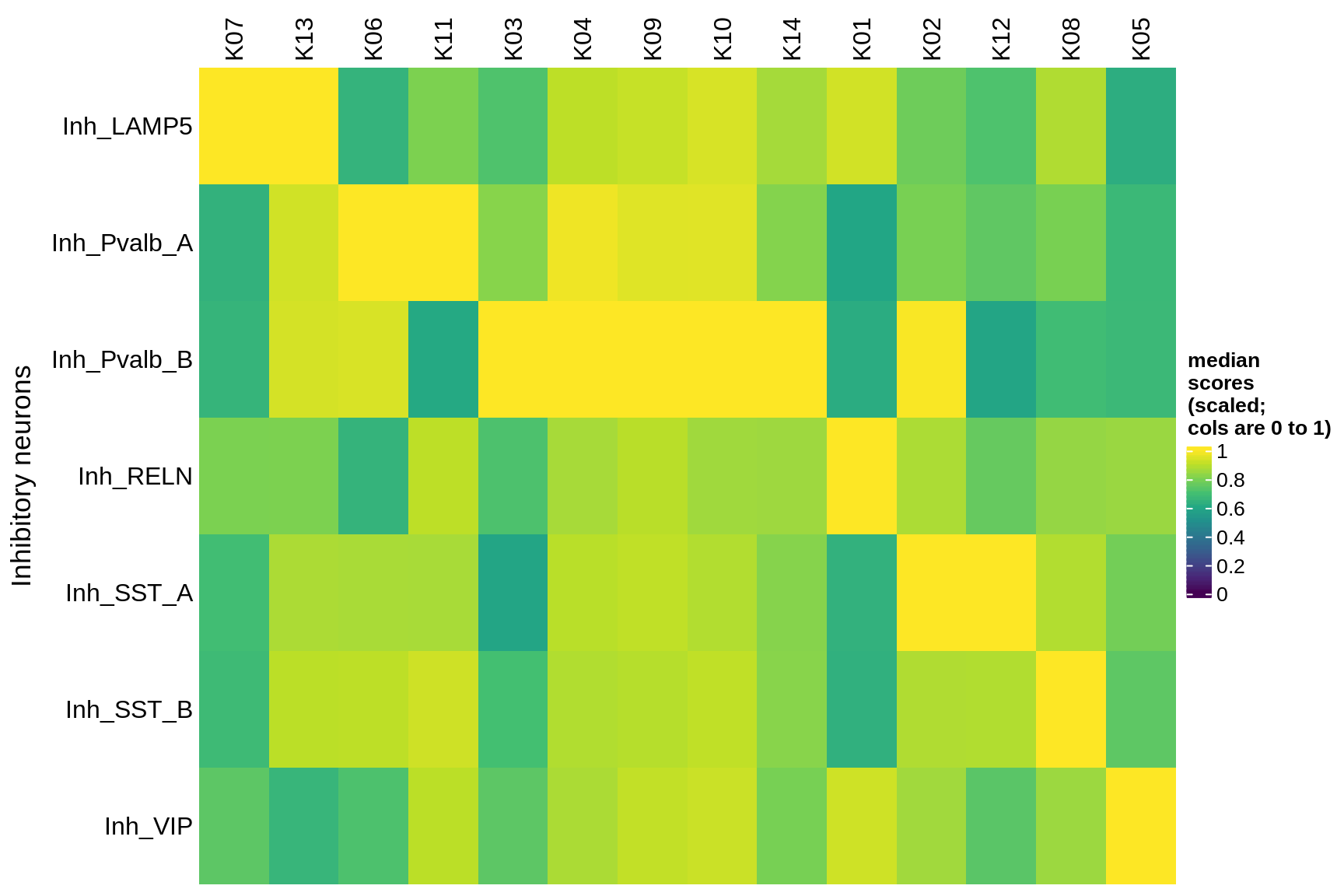
astrocytes
endo_stromal
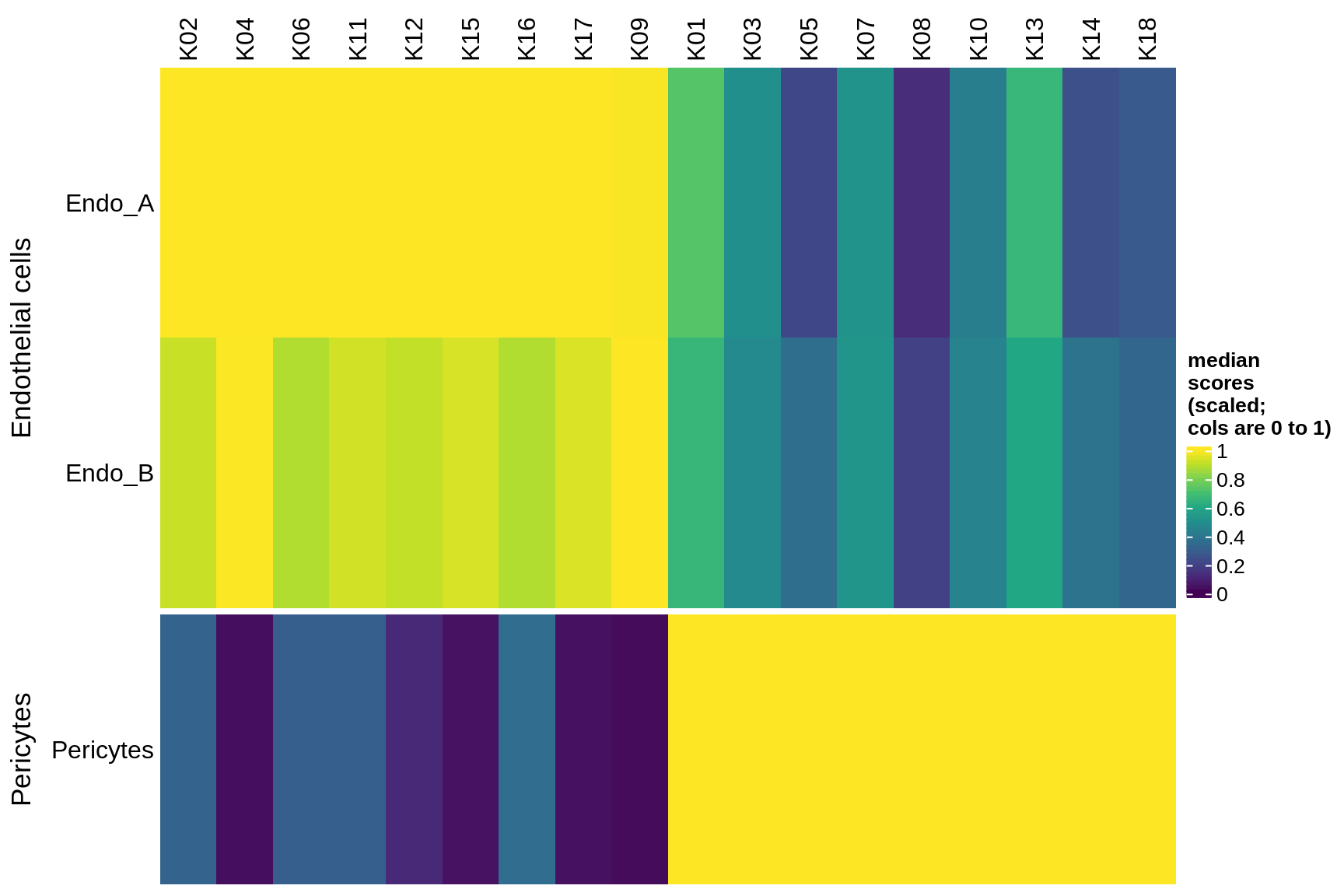
microglia
immune
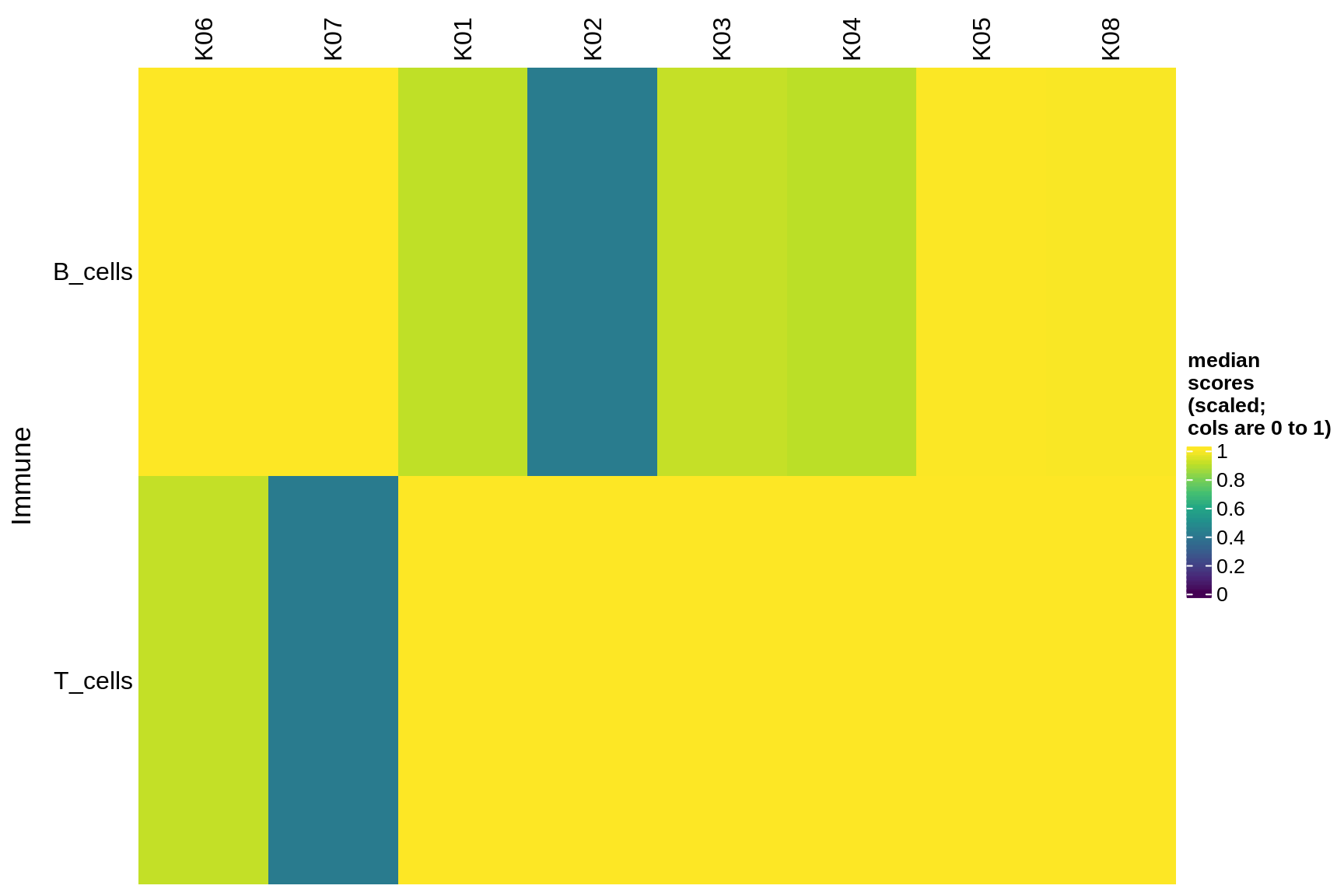
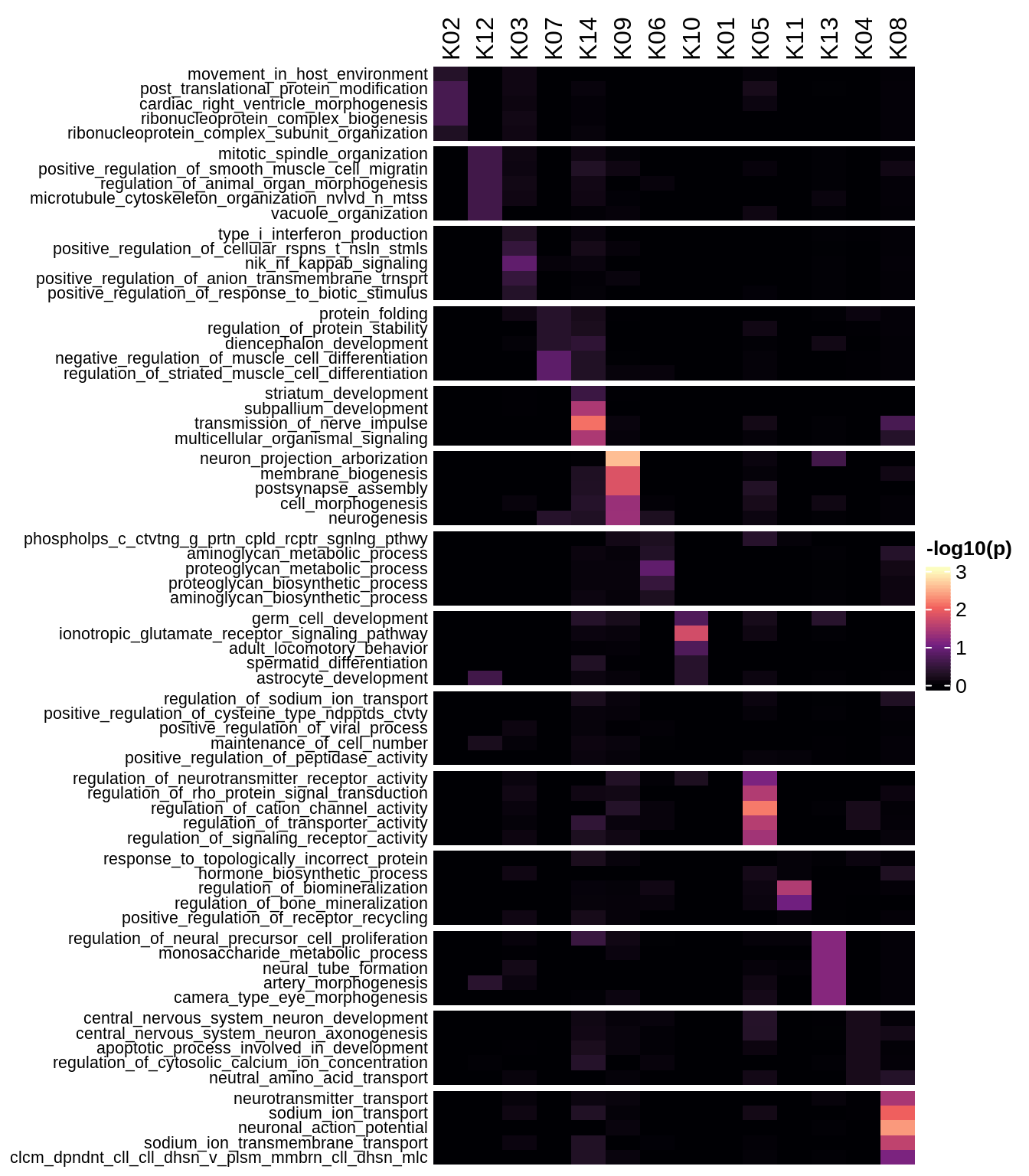
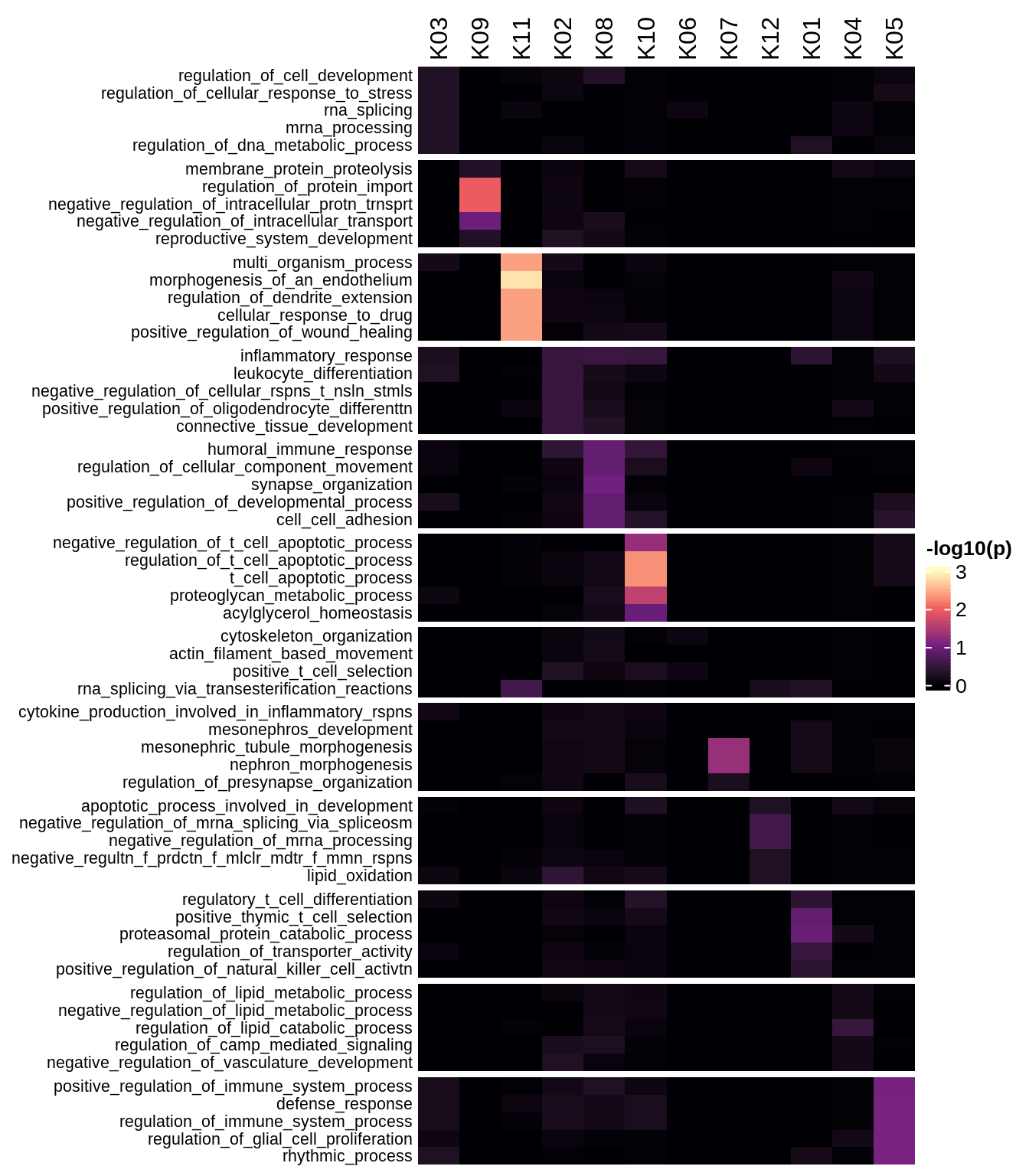
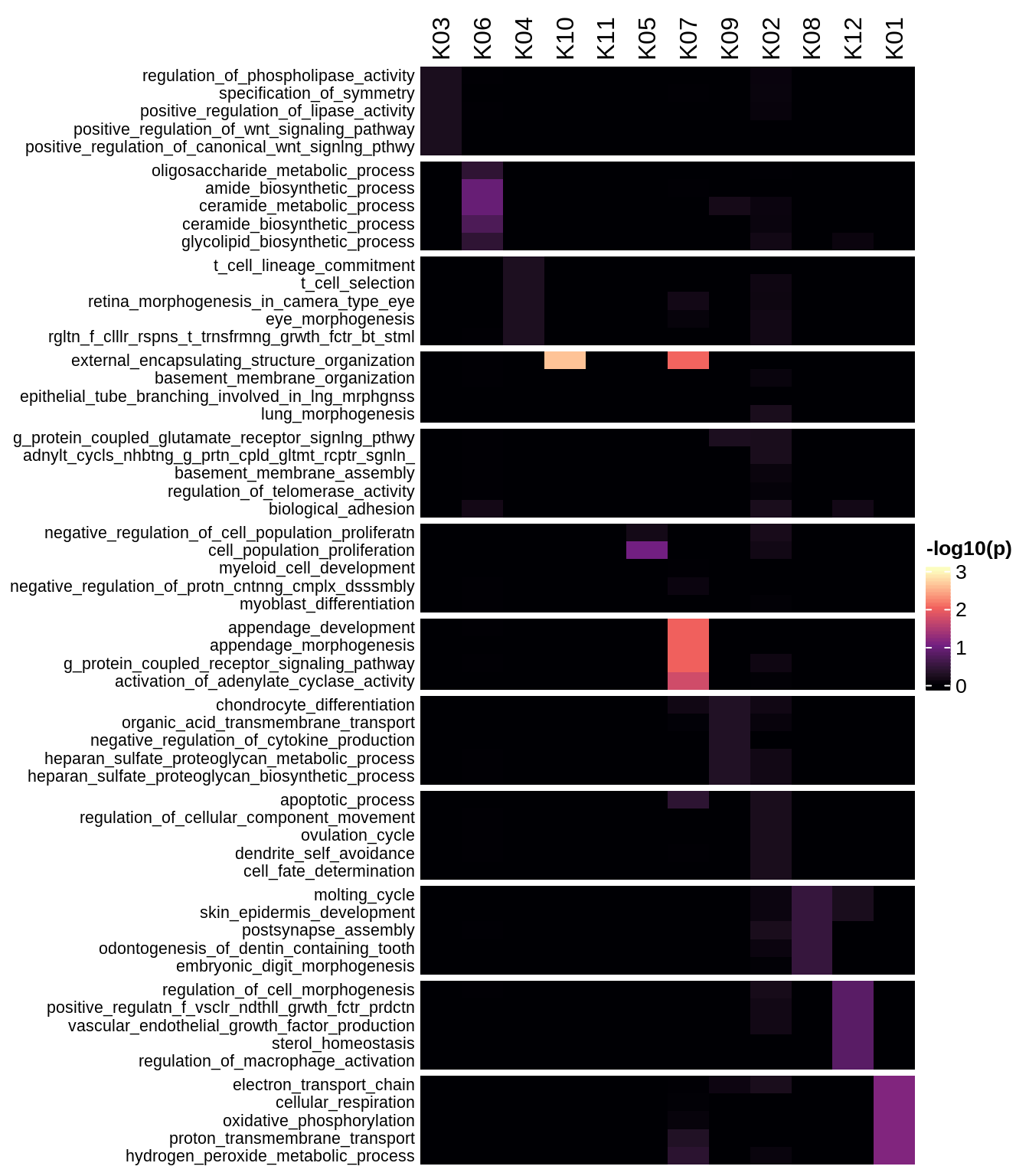
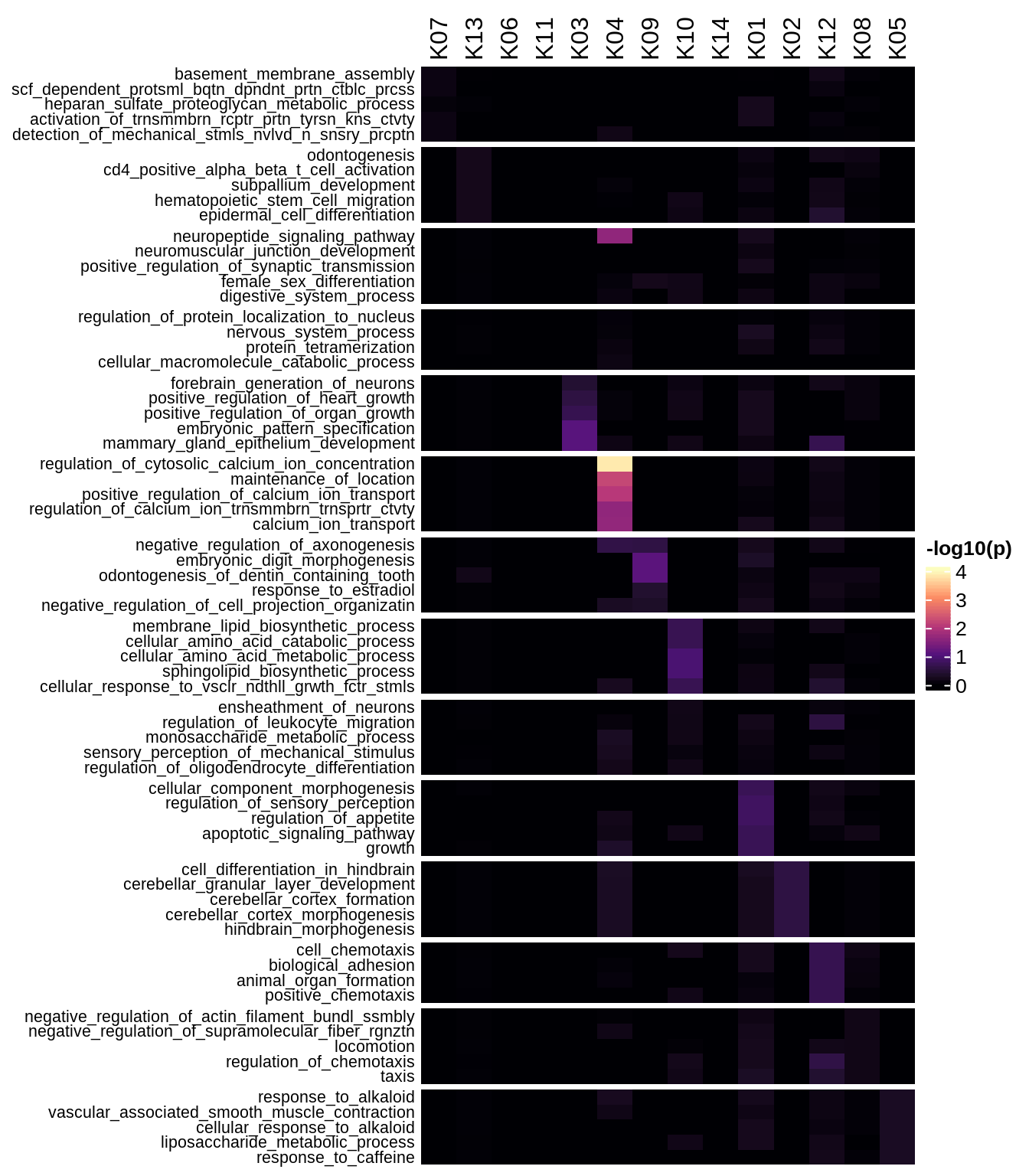
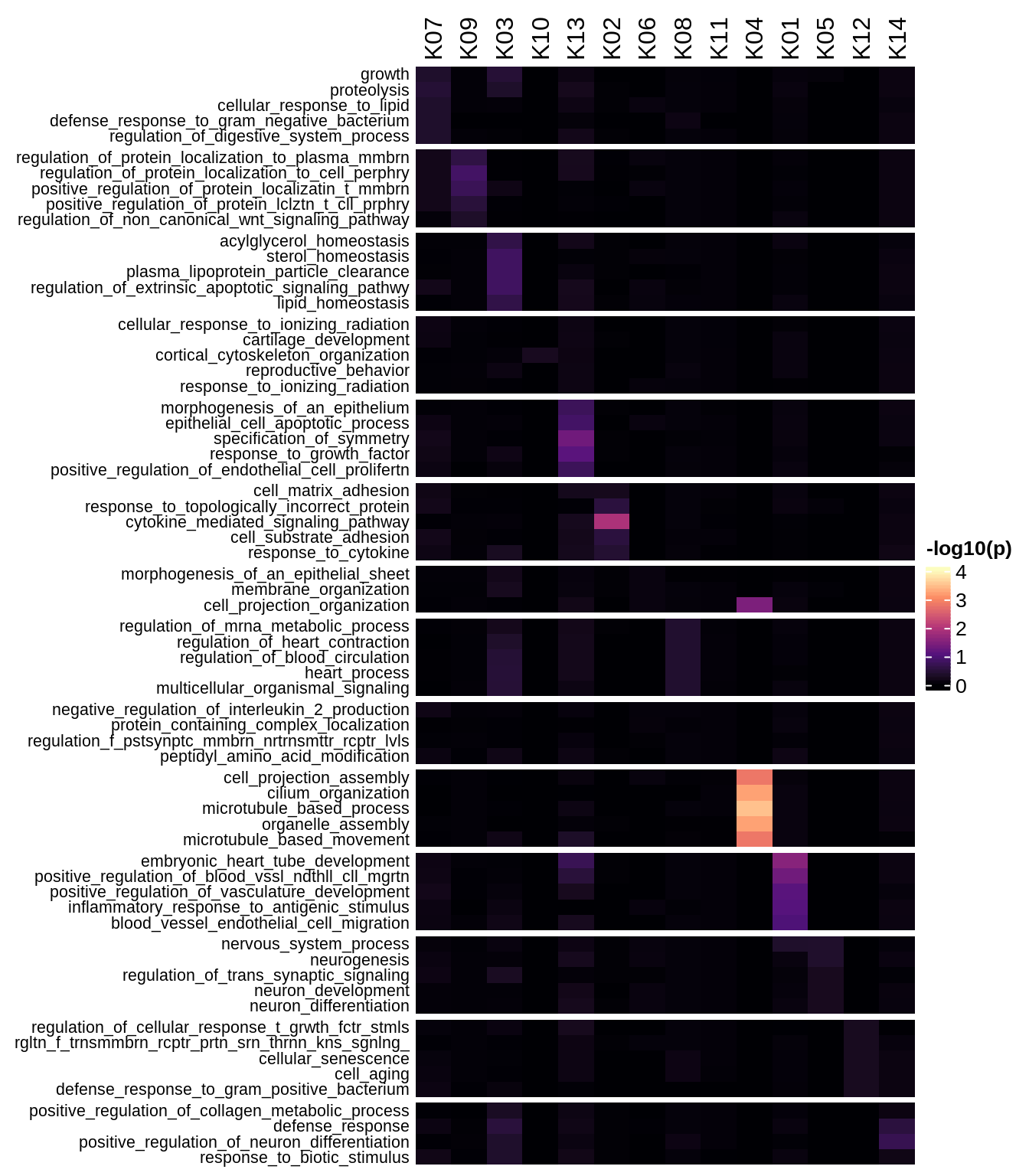
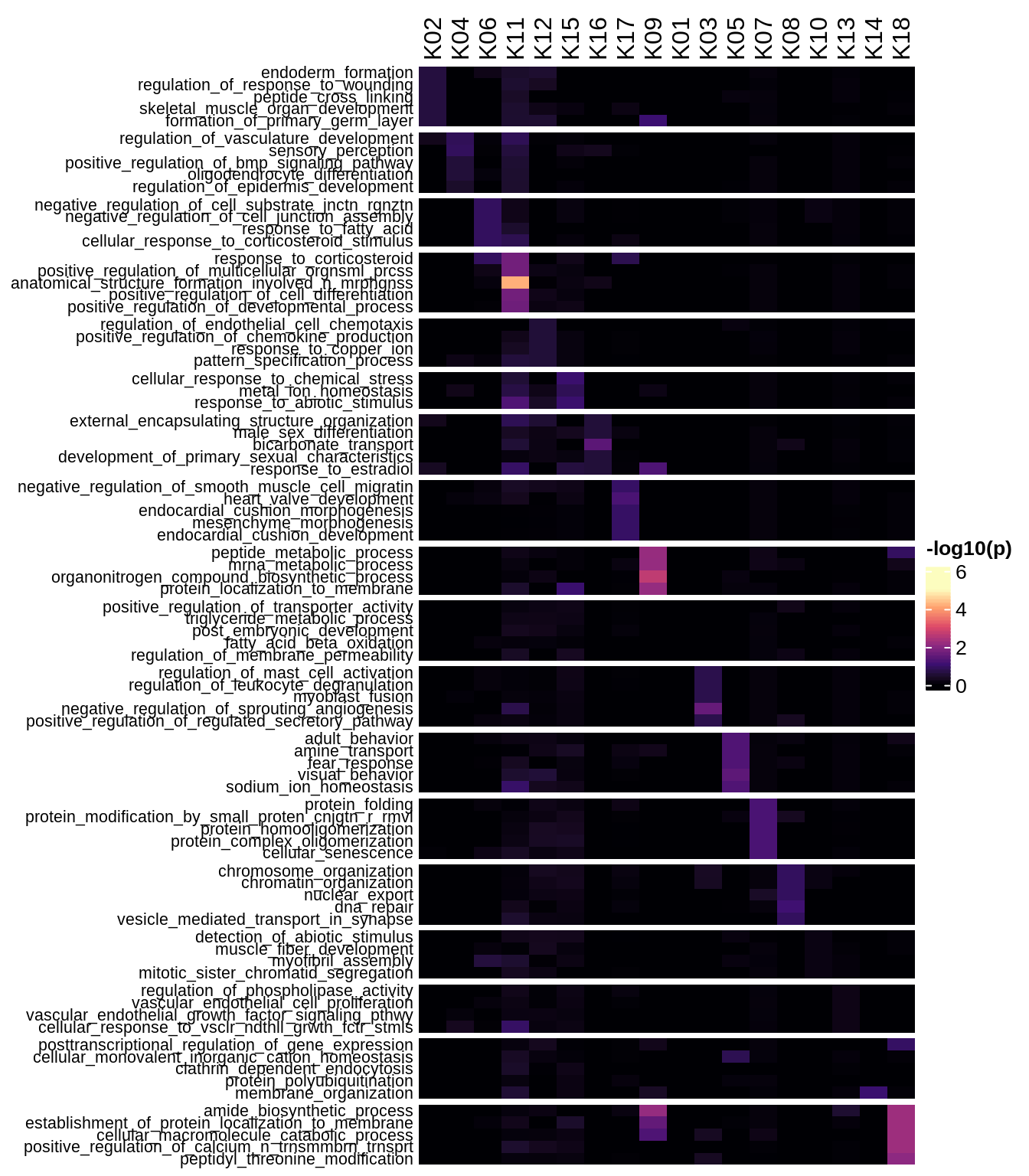
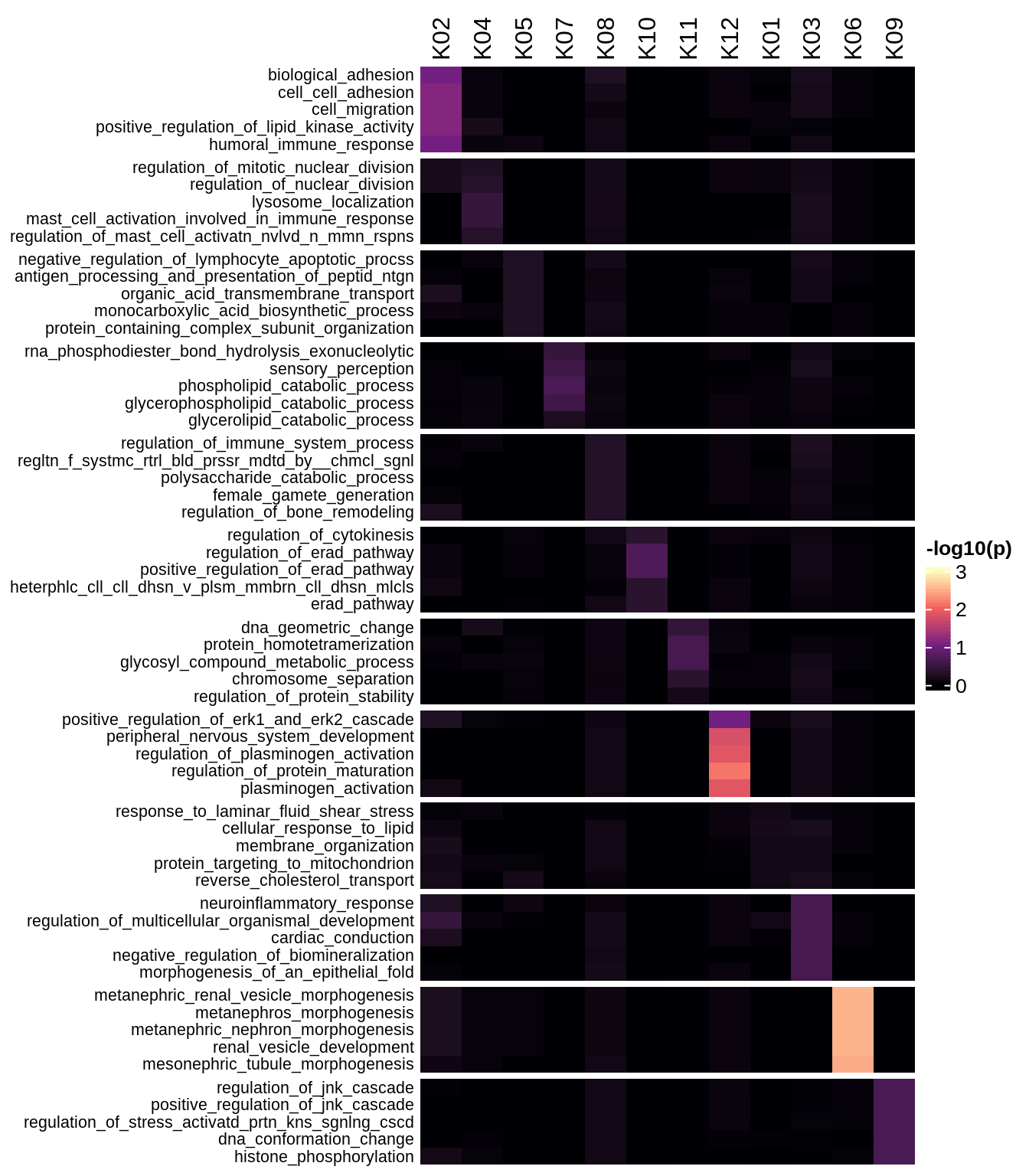
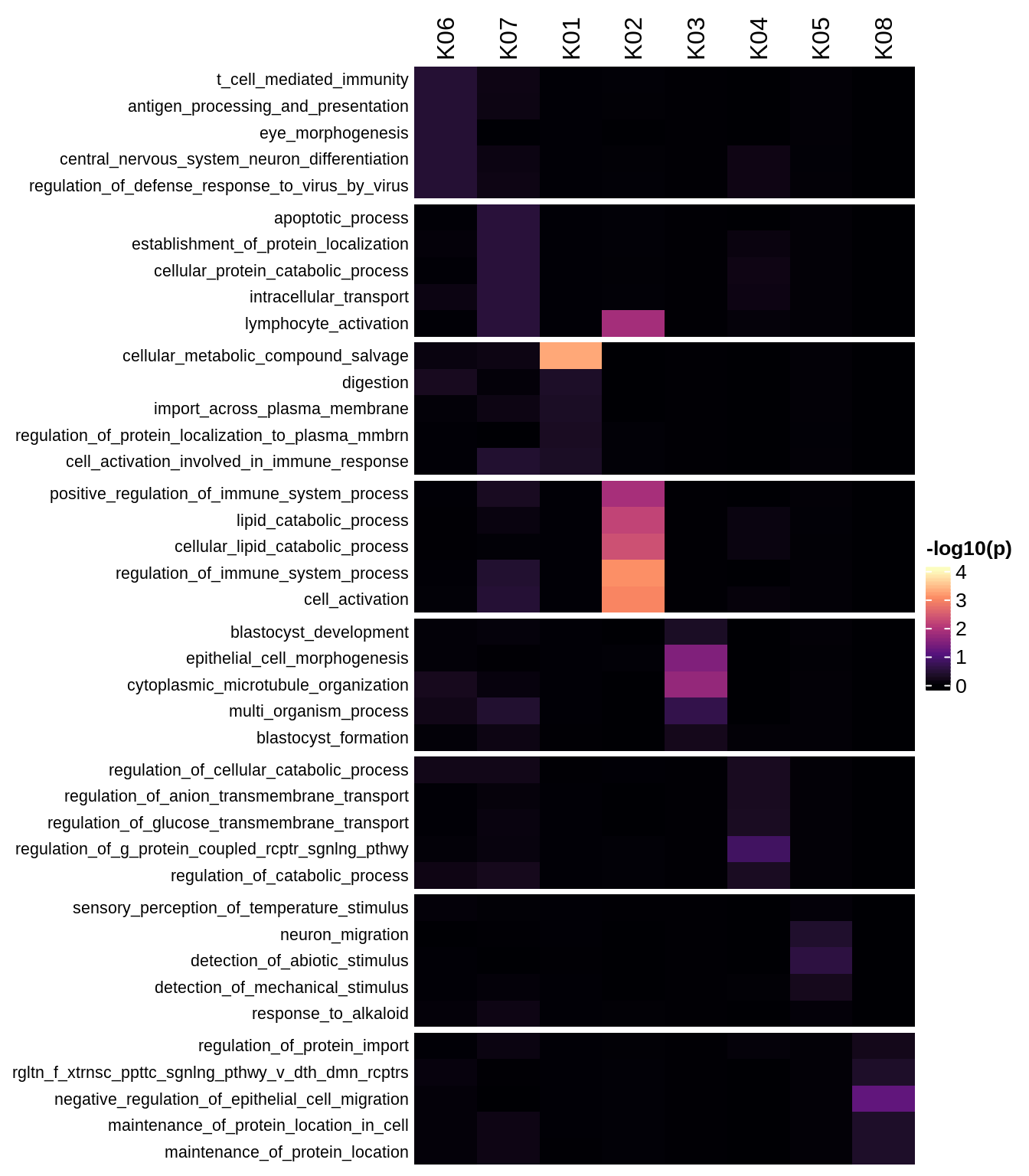
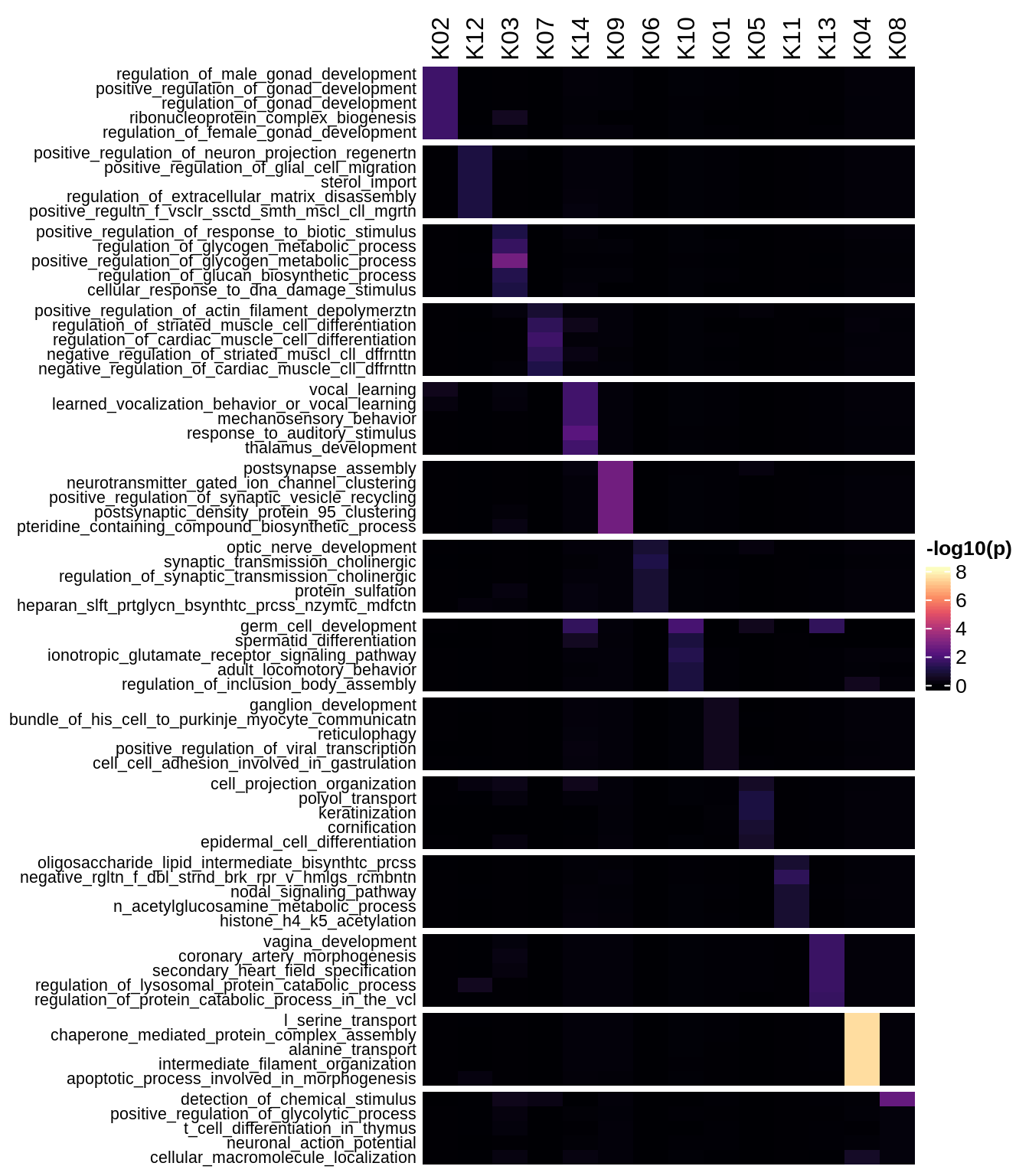
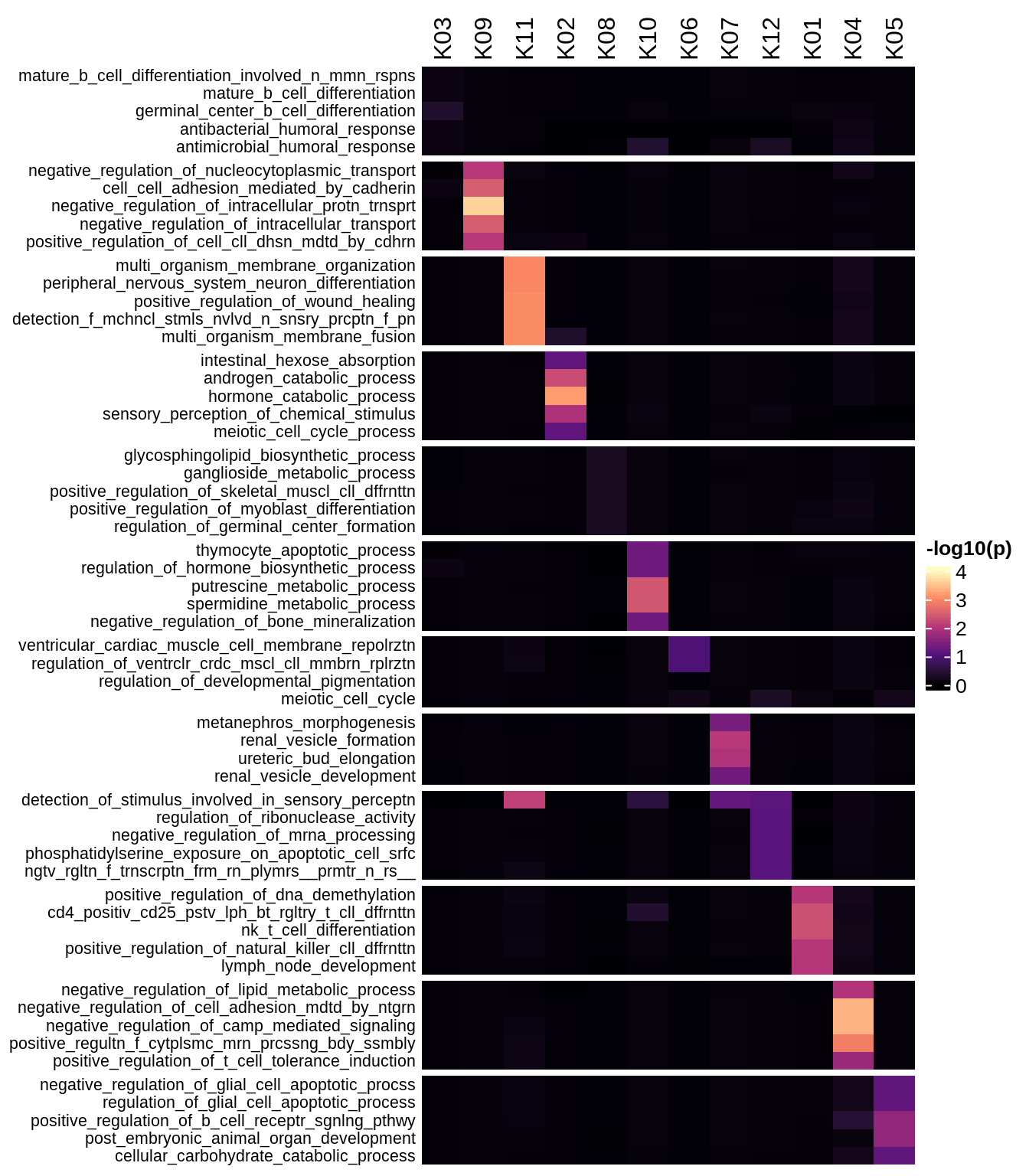
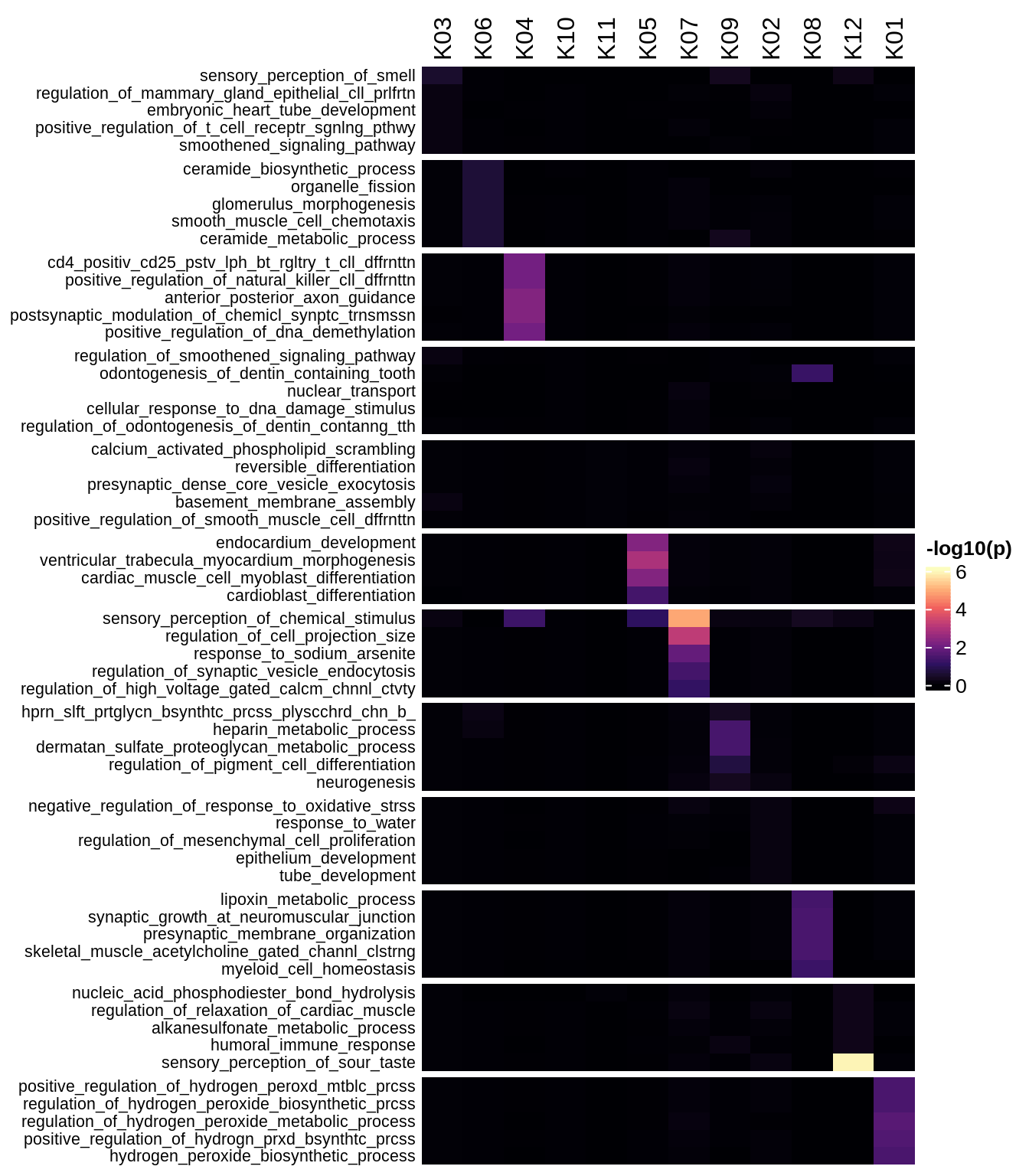
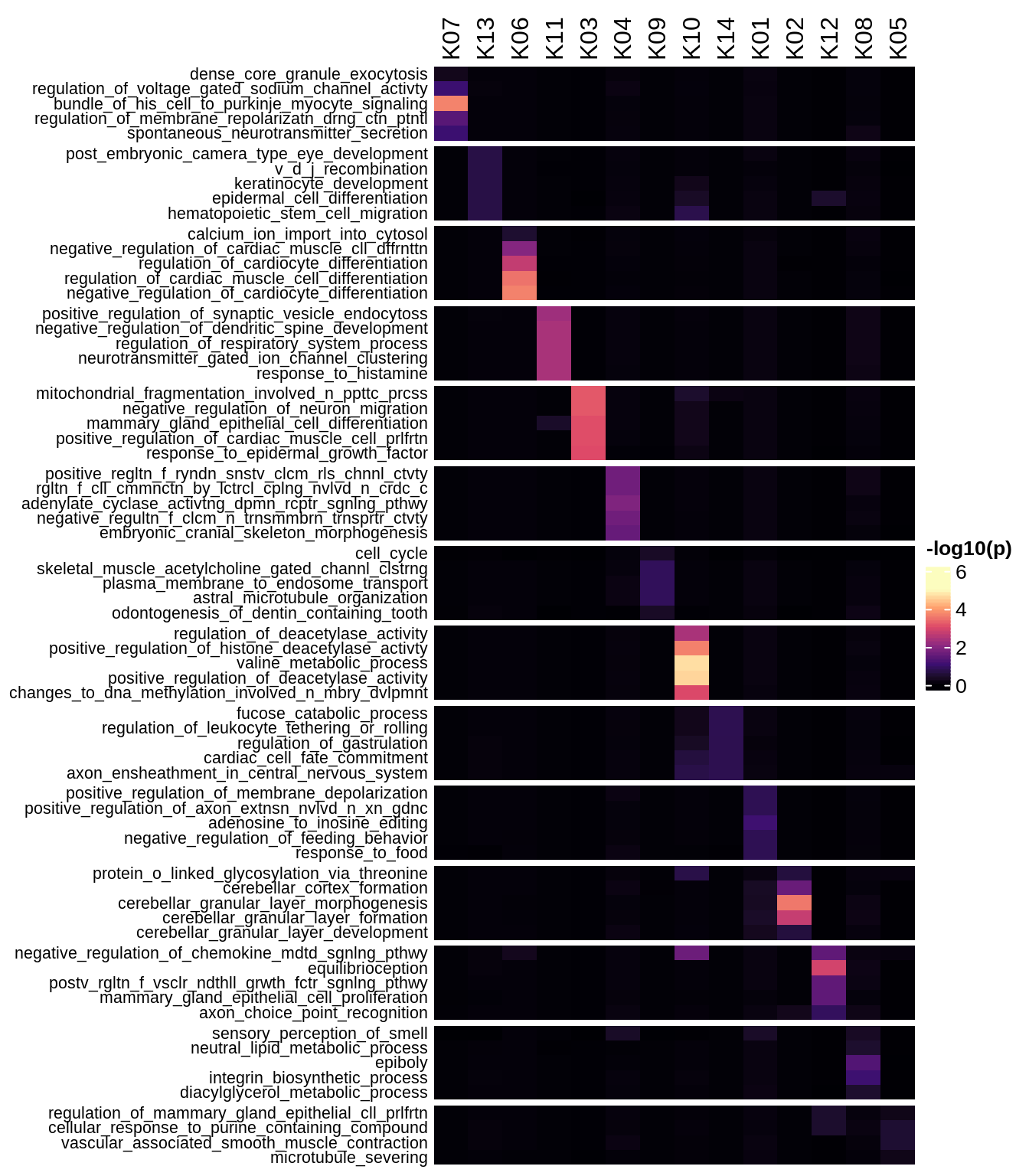
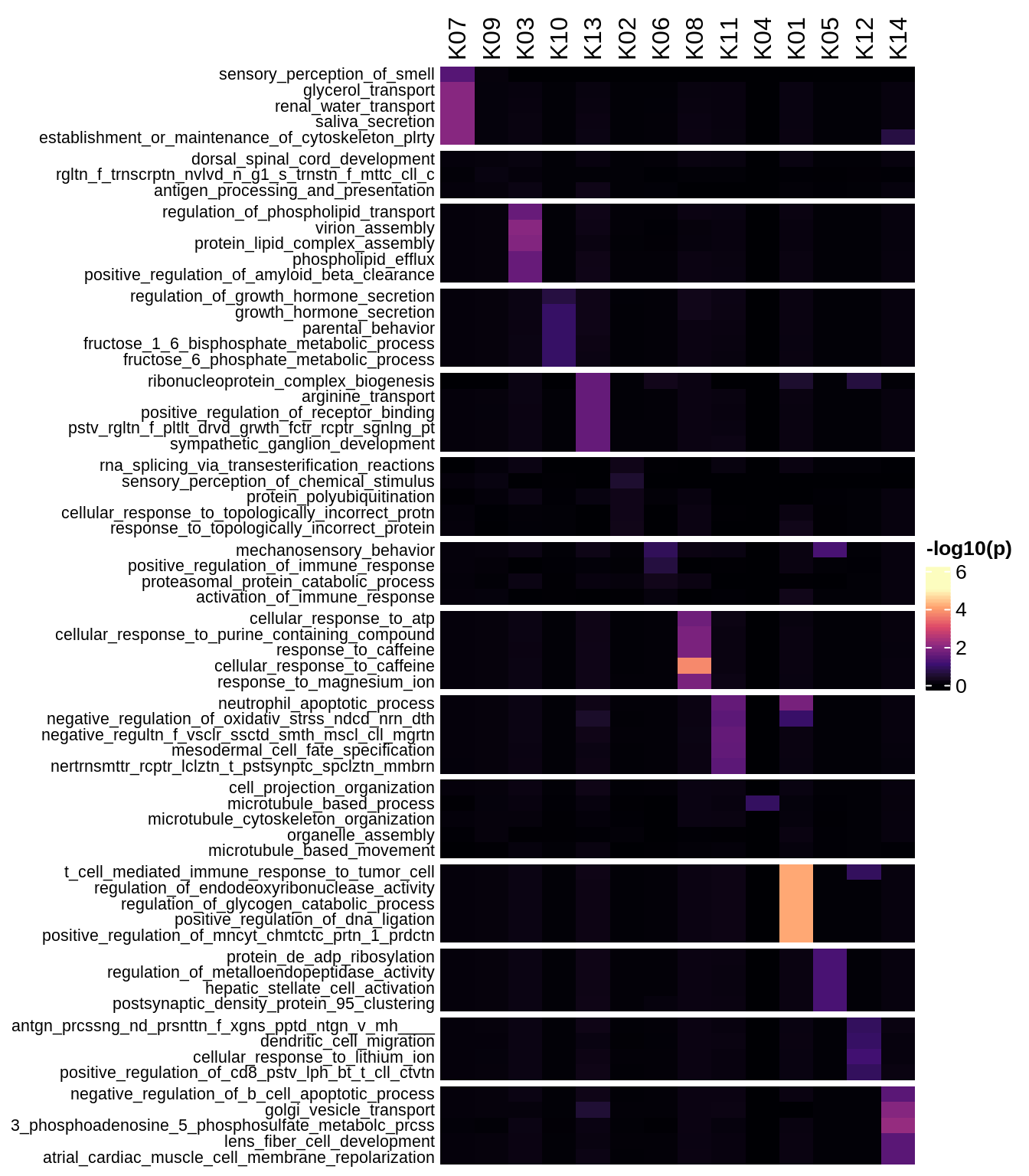
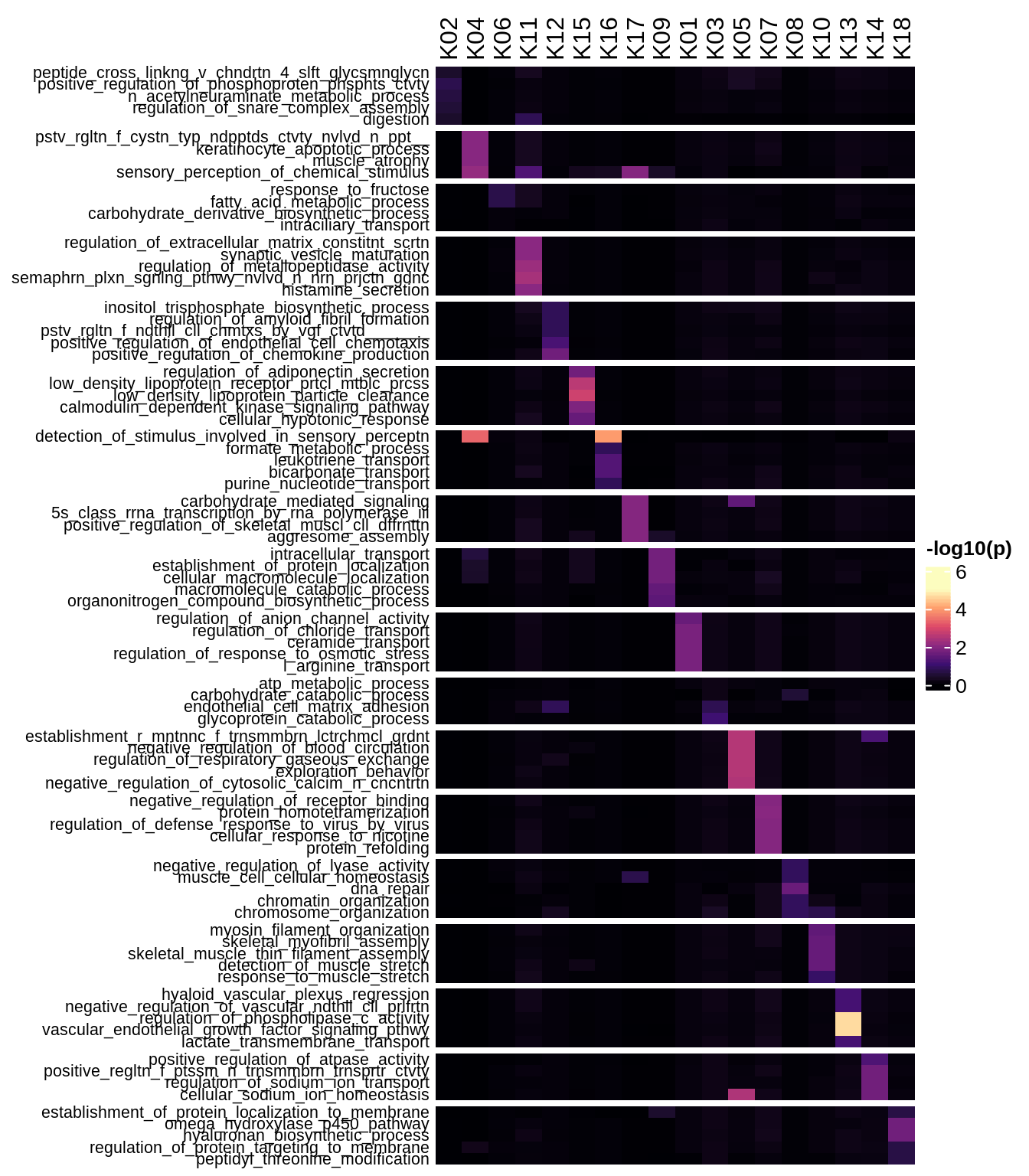
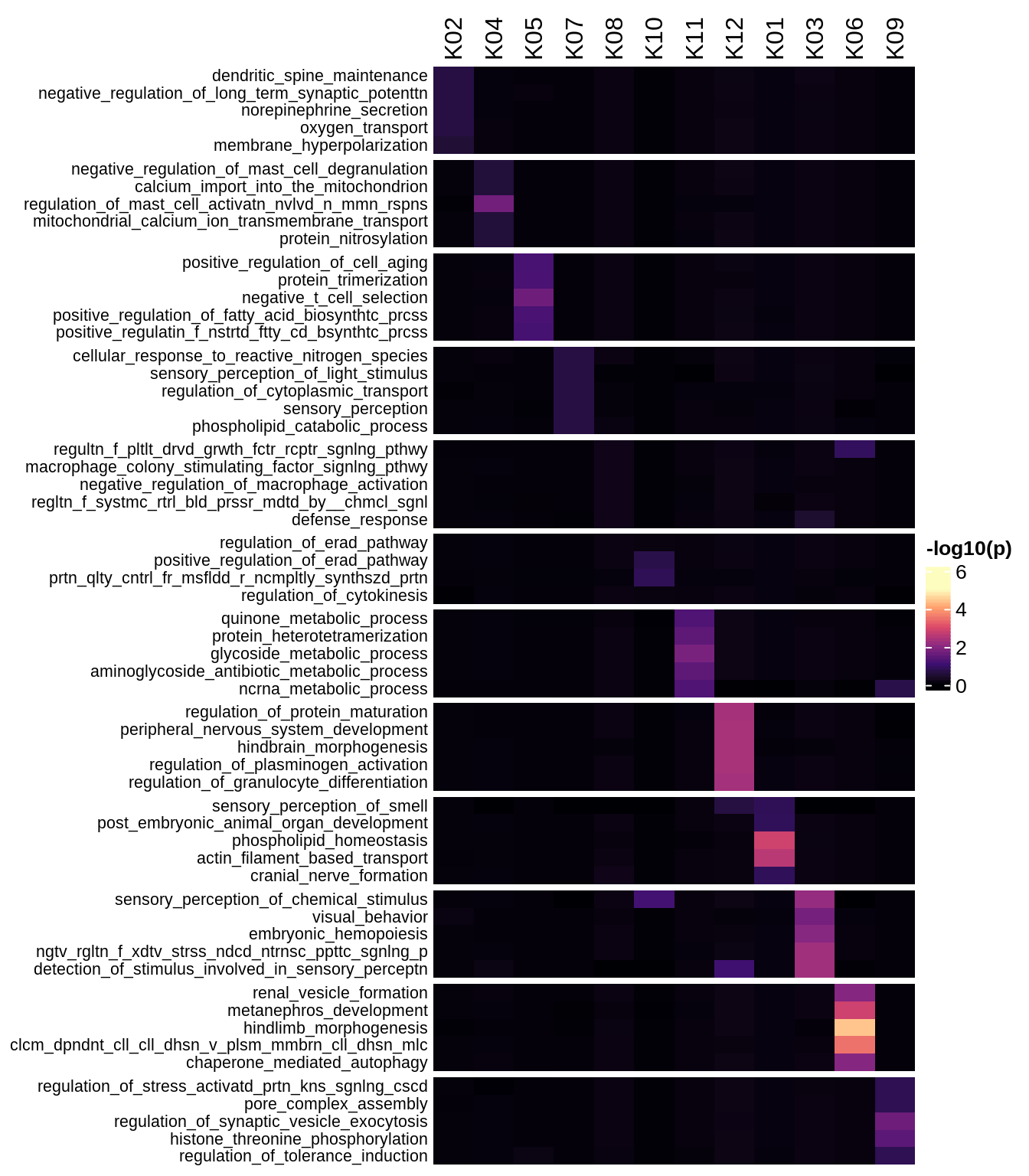
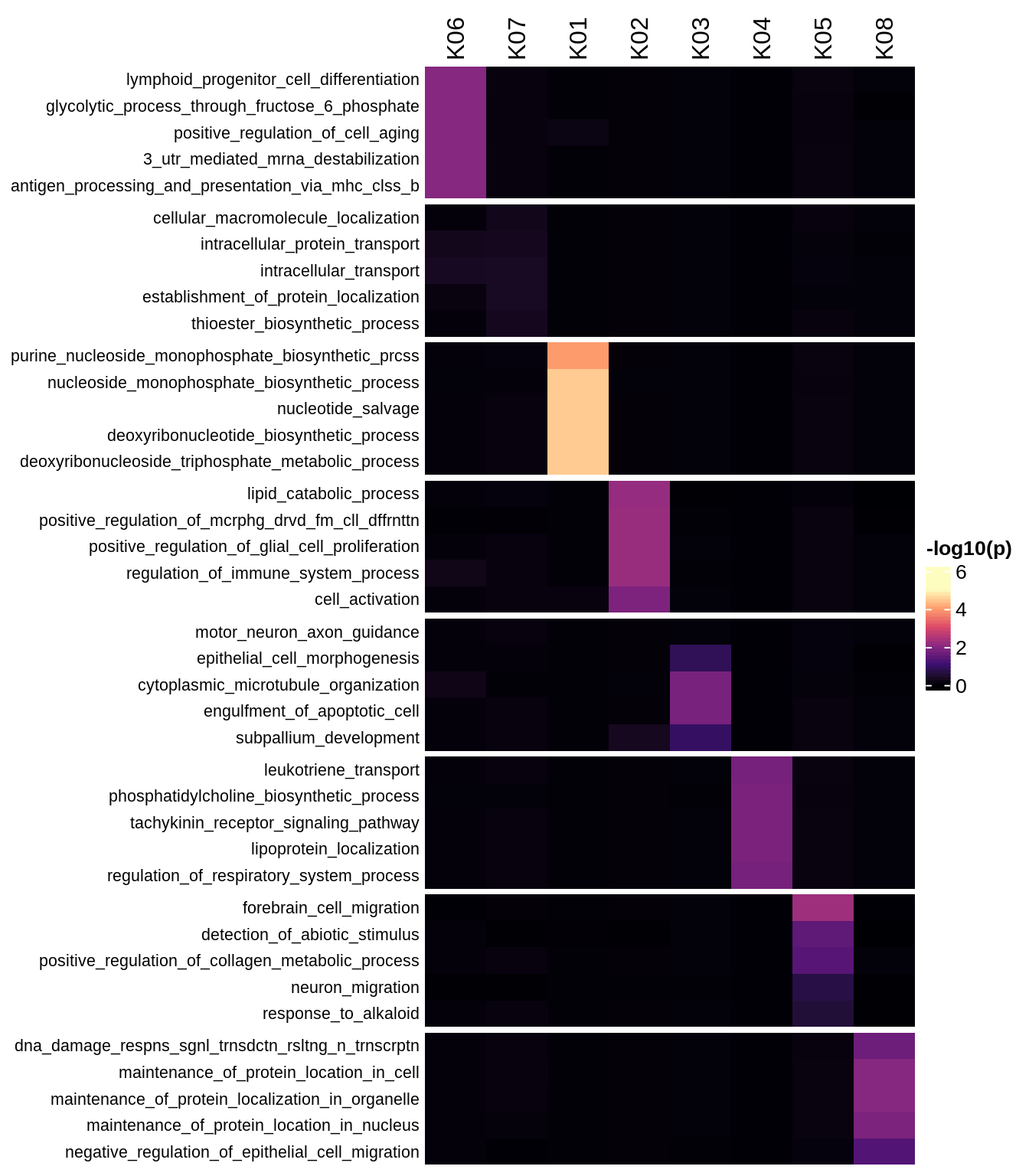
Enriched GO terms per module (using 2k genes for GSEA)
for (g in group_list) {
cat('### ', g, '\n')
hm = plot_enriched_sets(pop_list[[g]]$go_std_dt, pop_list[[g]]$k_order)
if (!is.null(hm))
draw(hm)
cat('\n\n')
}Enriched GO terms per module (using more genes for GSEA)
for (g in group_list) {
cat('### ', g, '\n')
hm = plot_enriched_sets(pop_list[[g]]$go_all_dt, pop_list[[g]]$k_order)
if (!is.null(hm))
draw(hm)
cat('\n\n')
}UMAP celltype reference
(plot_umap_celltypes(umap_dt, conos_dt))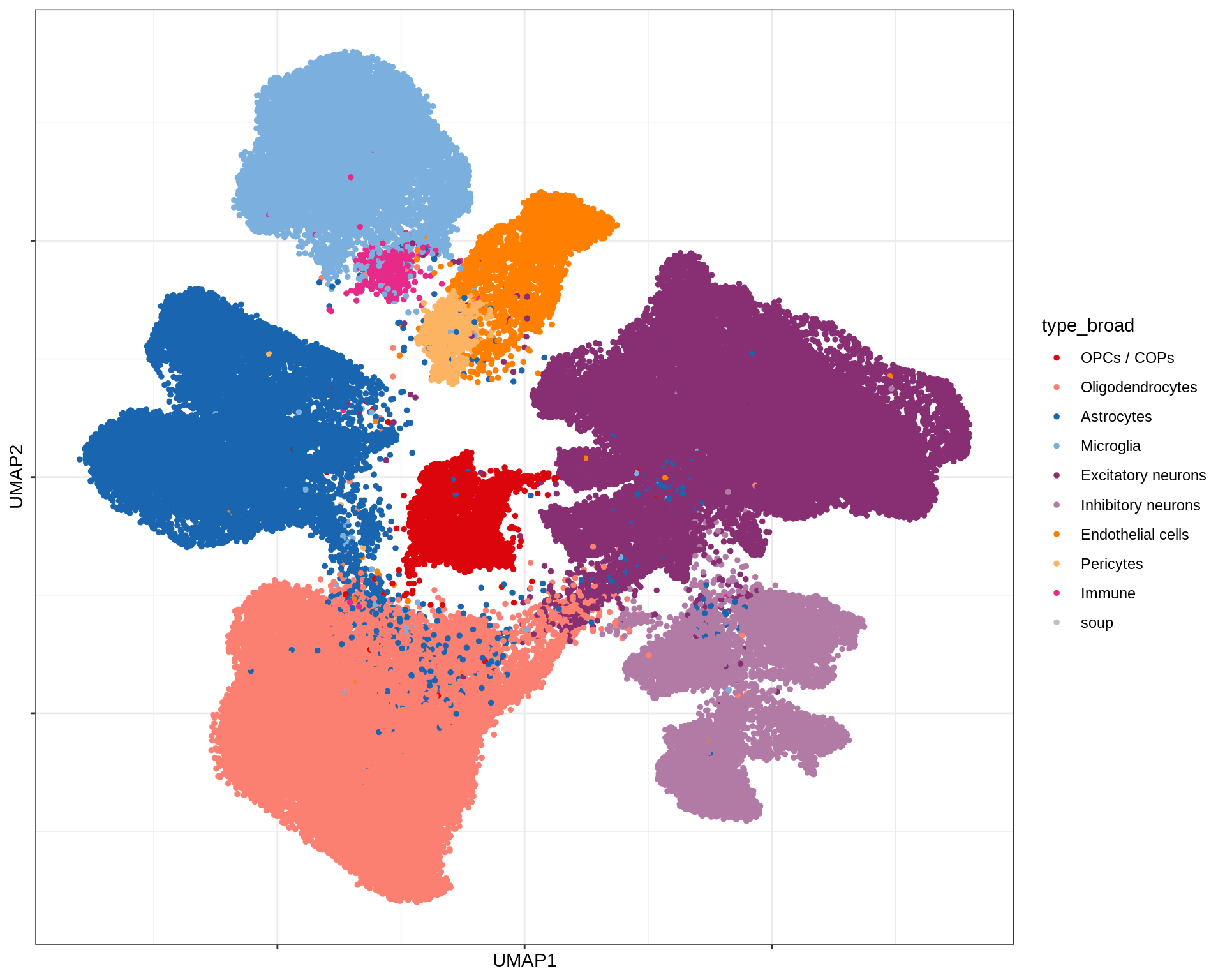
feat_list = scores_dt$feat %>% unique %>% sort %>% .[k_order]
for (f in feat_list) {
cat('### ', f, '\n')
print(plot_scores_over_lesions(scores_dt, f, meta_dt))
cat('\n\n')
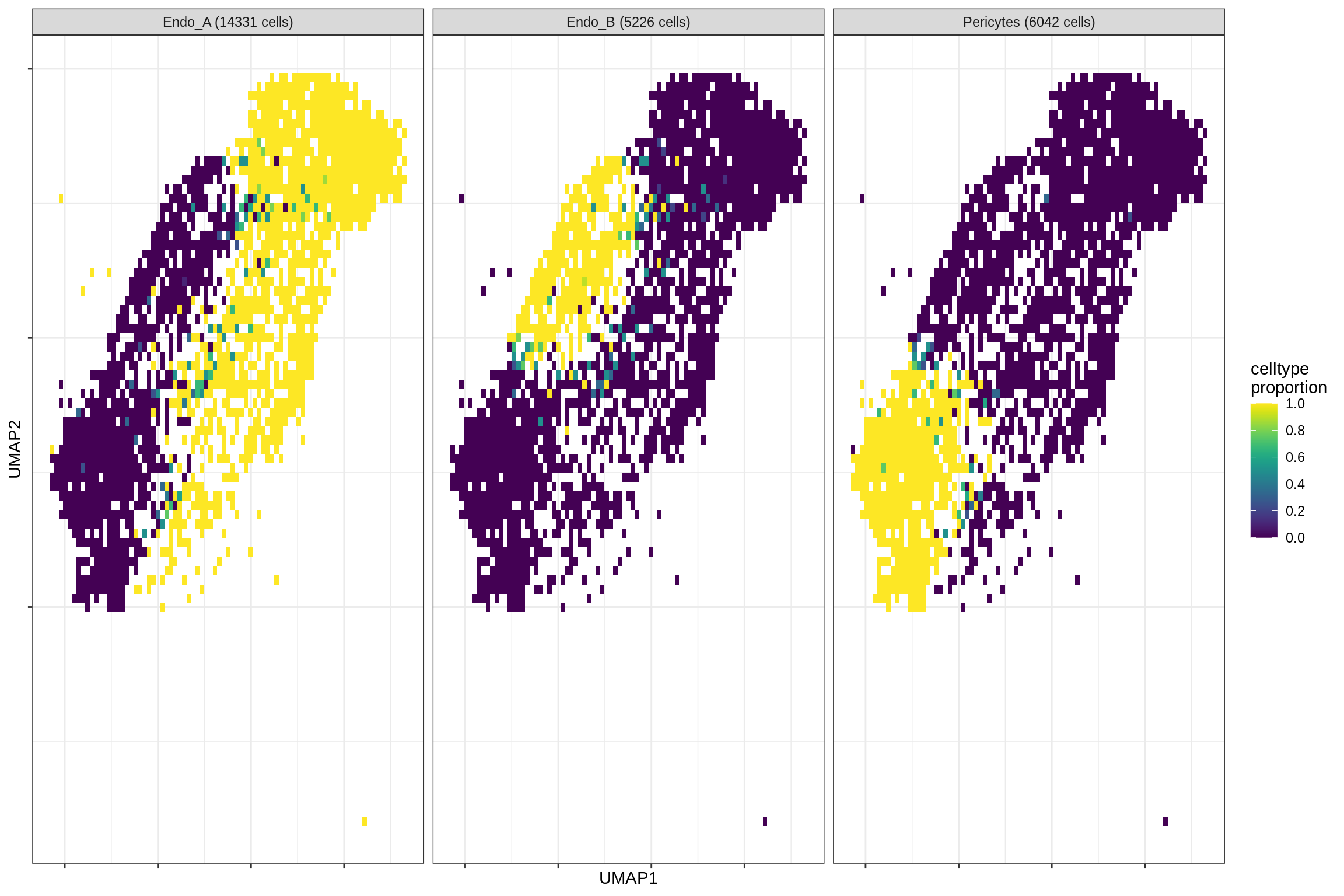
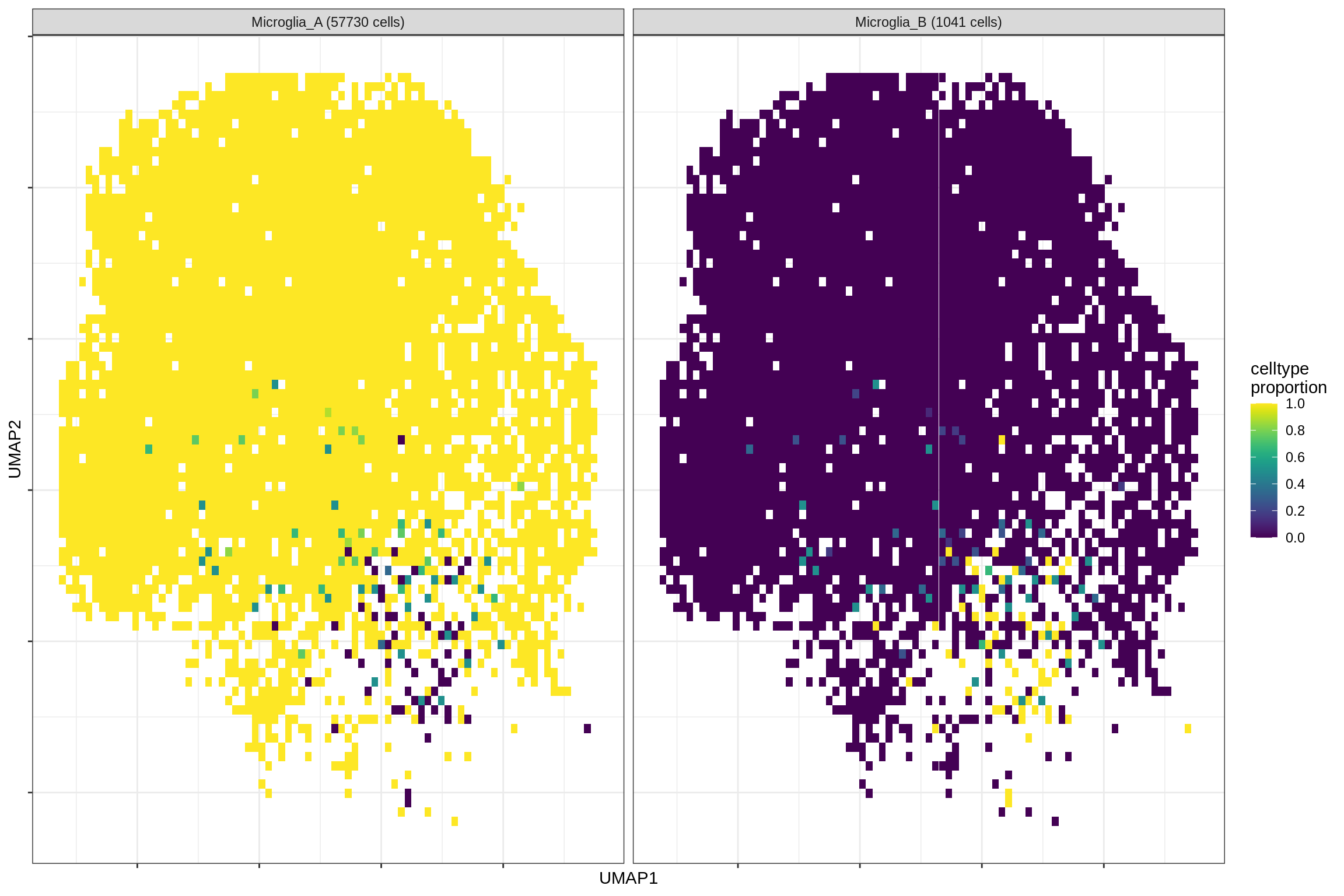
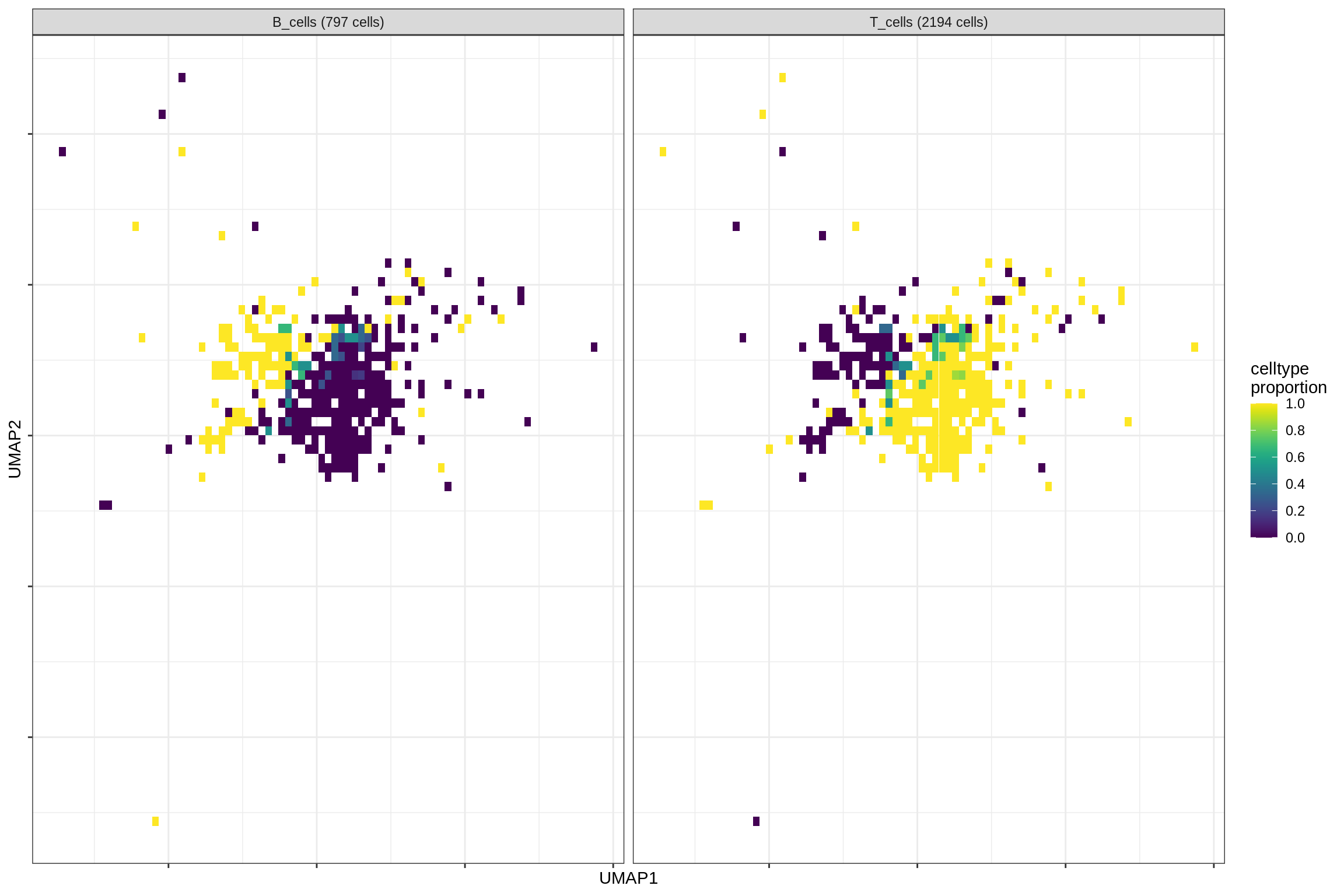
}Distribution of fine celltypes over UMAP
for (g in group_list) {
cat('### ', g, '\n')
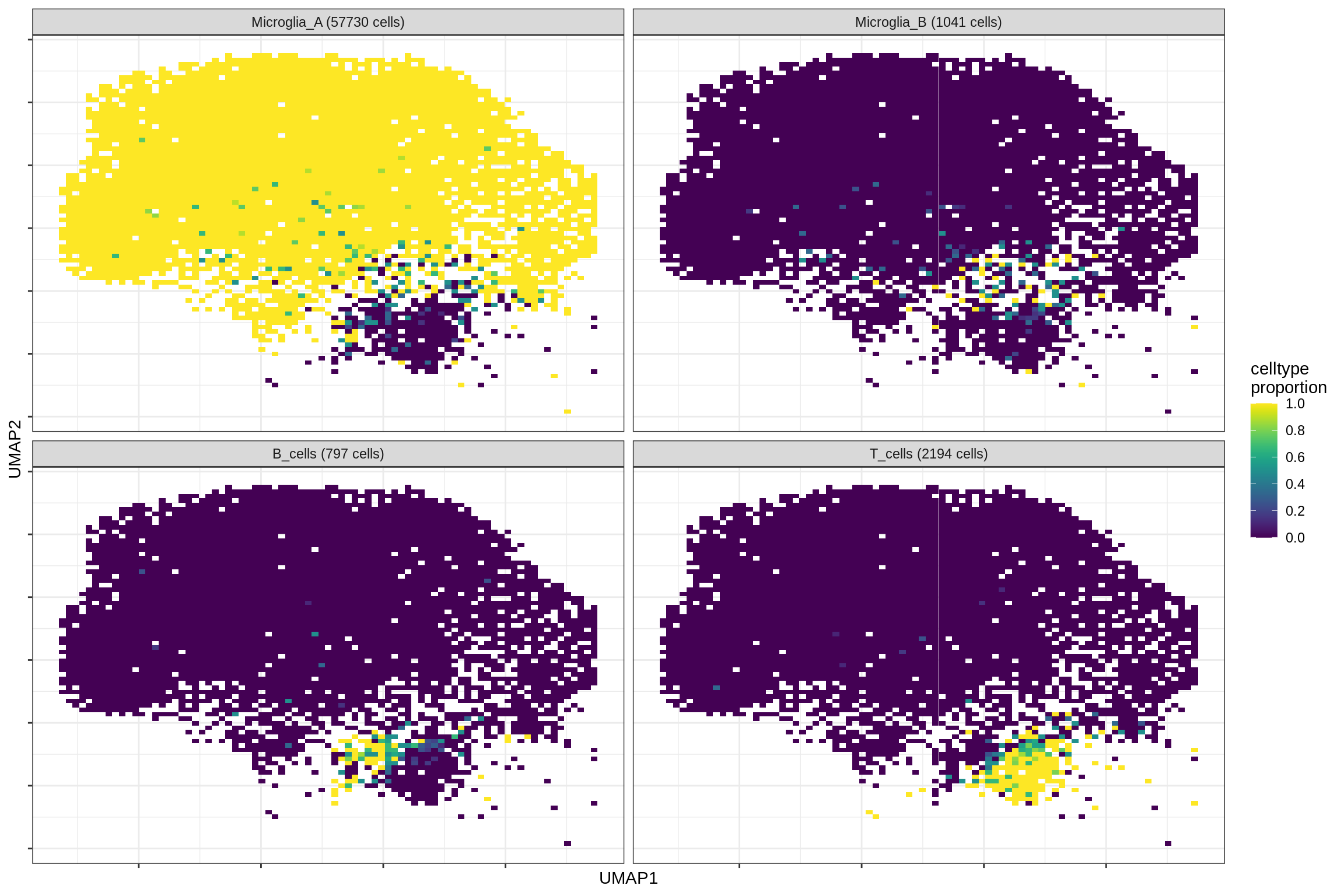
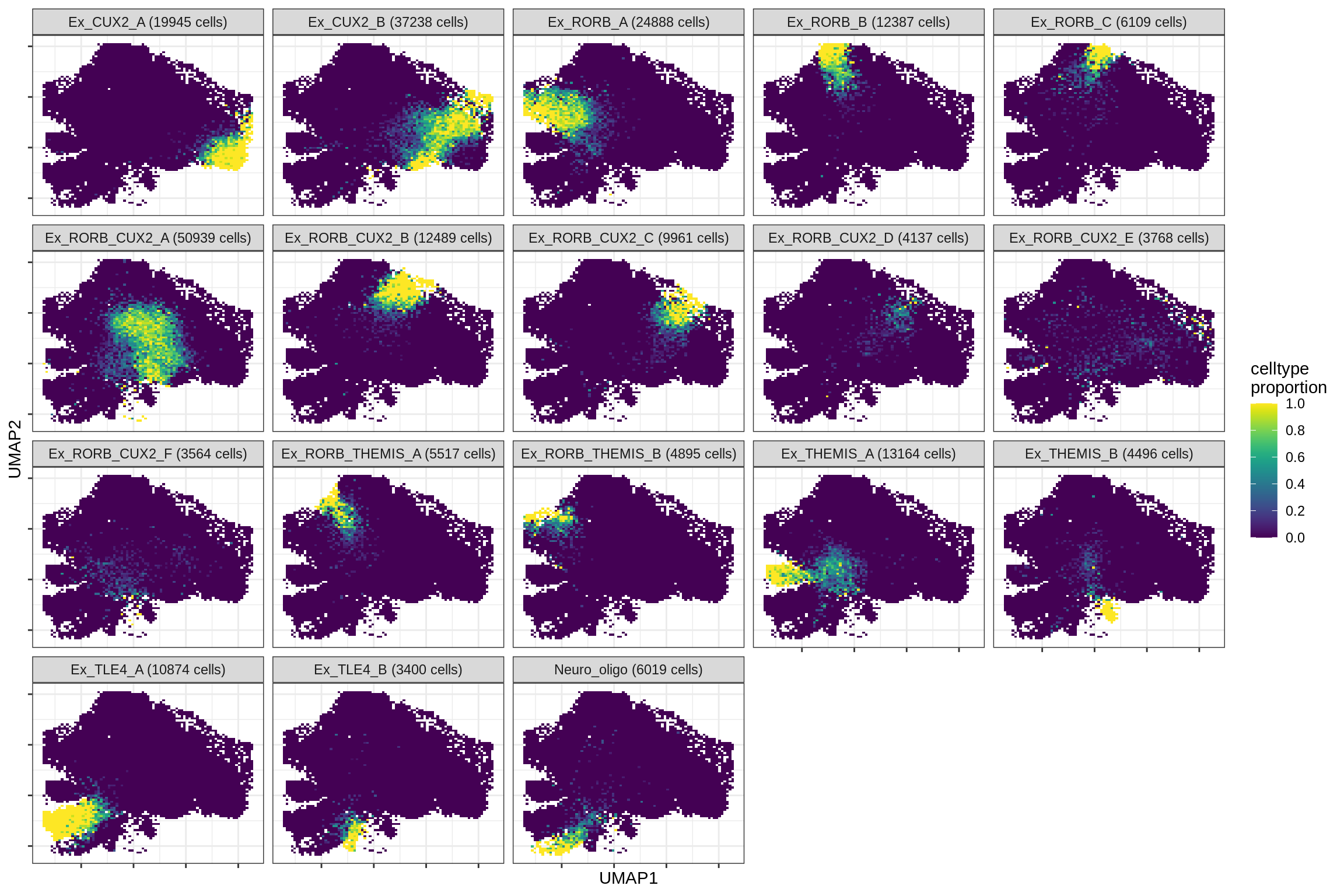
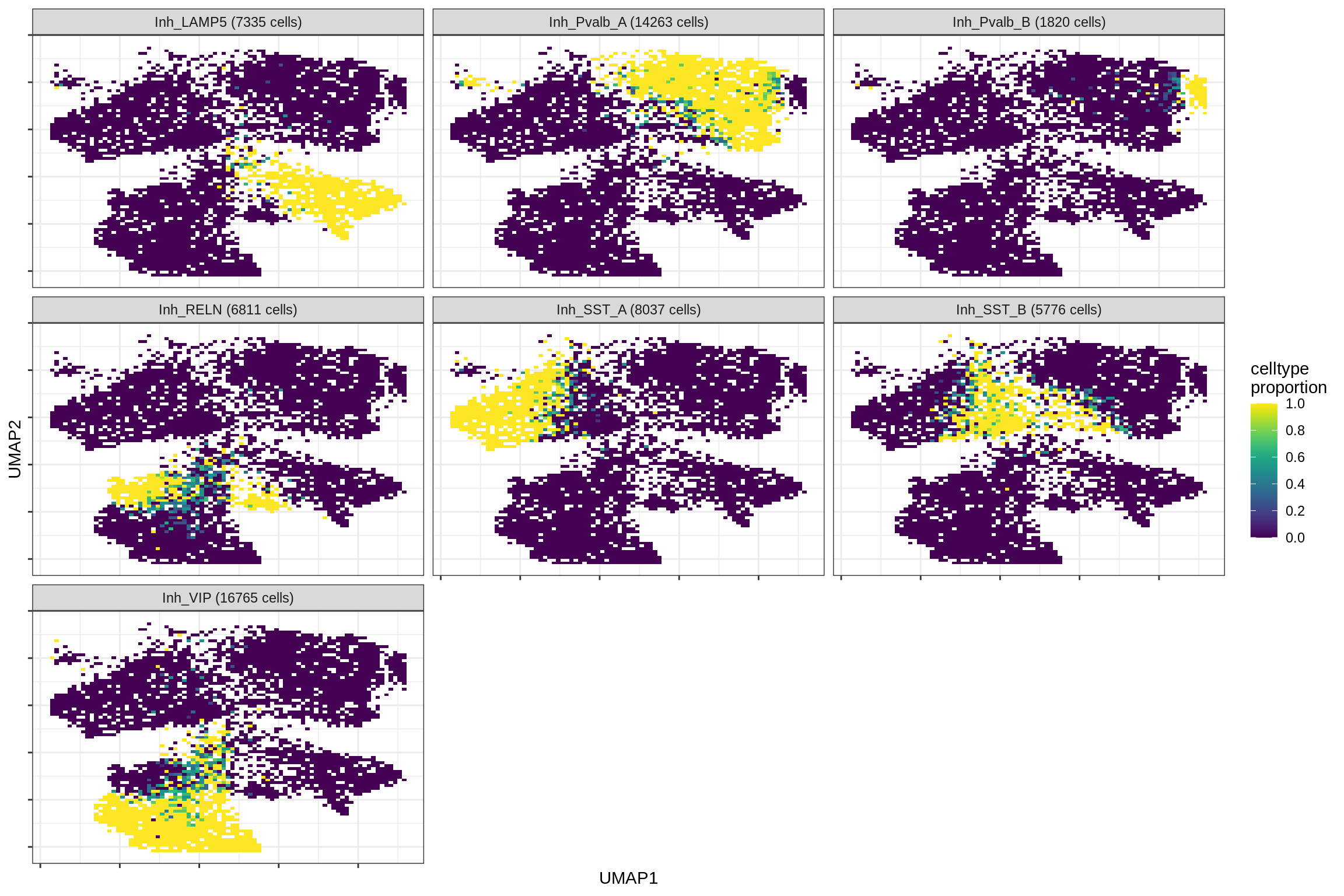
print(plot_celltypes_over_umap(spec_list[[g]], conos_dt, umap_dt))
cat('\n\n')
}oligo_opc
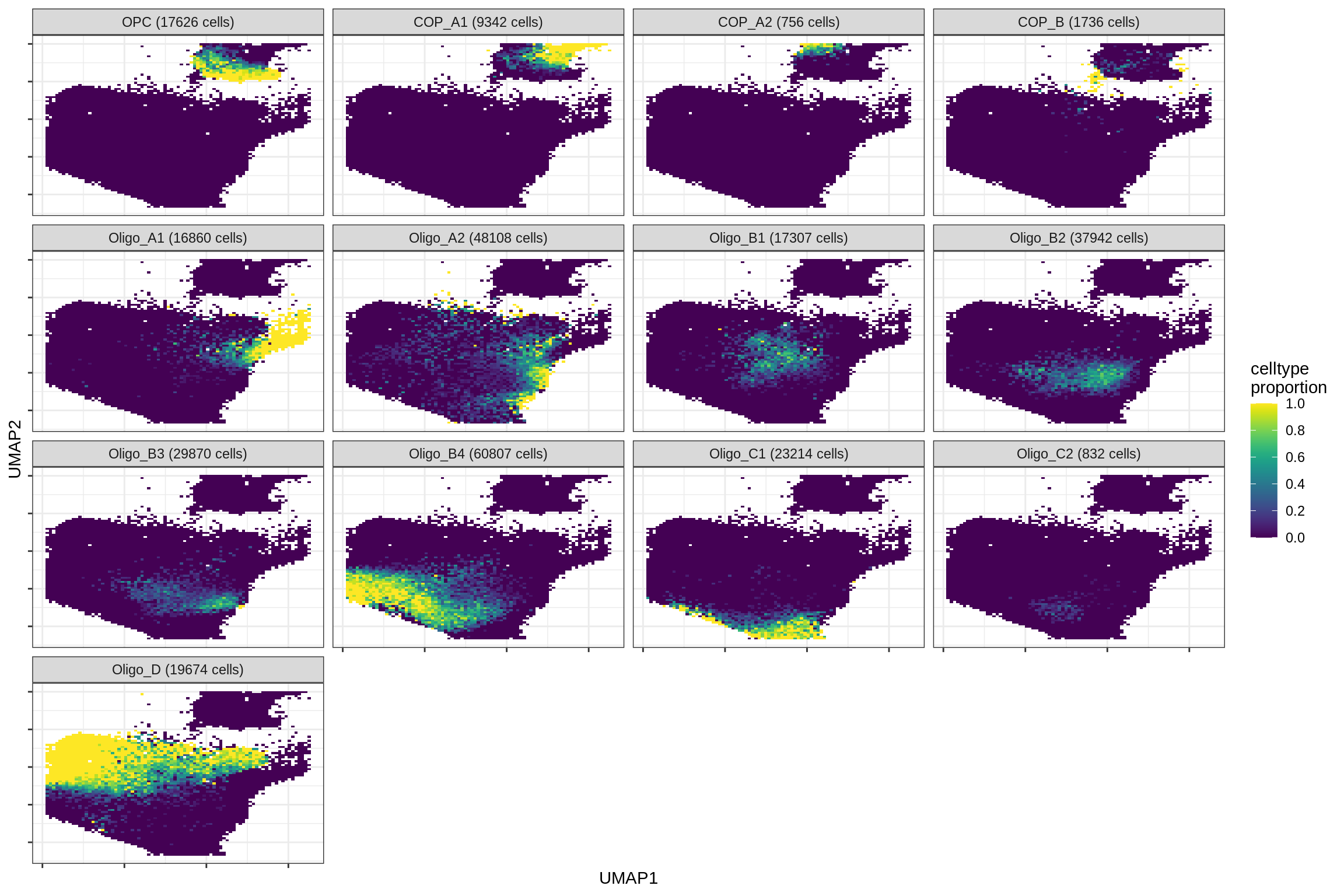
micro_immune
excitatory
inhibitory
astrocytes
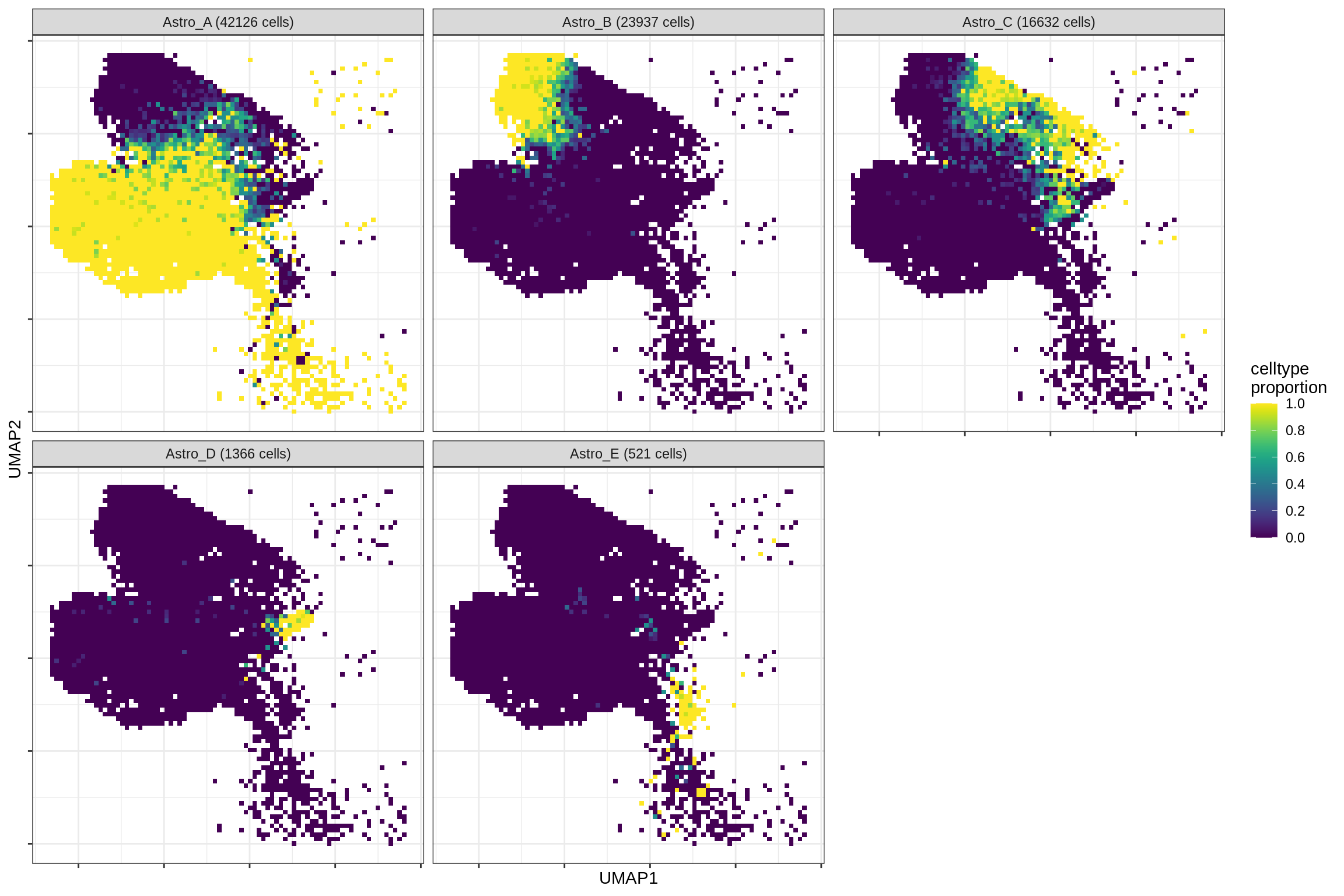
endo_stromal
microglia
immune
Outputs
save_module_genes_to_xl(pop_list, xl_f) broad_type symbol K01 K02 K03 K04
1: oligo_opc PLP1 2.531141e-02 2.532711e-14 1.029482e-14 2.037848e-02
2: oligo_opc DSCAM 2.274387e-18 6.132759e-02 1.197744e-02 1.965392e-18
3: oligo_opc CTNNA3 1.456599e-01 1.848812e-14 6.967282e-15 6.473713e-03
4: oligo_opc OPCML 2.274387e-18 1.852172e-02 4.558979e-02 1.965392e-18
5: oligo_opc ST18 1.490506e-01 1.861083e-14 7.252551e-15 8.745914e-03
---
1996: oligo_opc ARL4A 2.274387e-18 7.160648e-10 5.156852e-11 1.965392e-18
1997: oligo_opc SLC25A25 2.274387e-18 4.743476e-11 2.610729e-02 1.965392e-18
1998: oligo_opc NEDD4 9.946457e-08 3.733592e-02 1.627897e-02 7.060745e-06
1999: oligo_opc PFKP 2.274387e-18 1.553894e-09 2.652903e-02 1.965392e-18
2000: oligo_opc DEF8 5.791248e-09 2.552419e-14 1.150755e-14 3.035459e-02
K05 K06 K07 K08 K09
1: 3.723254e-02 2.428265e-02 2.915411e-14 2.971436e-04 1.212193e-02
2: 1.036054e-16 3.954935e-16 1.626423e-01 1.877257e-15 1.268171e-14
3: 3.980145e-02 7.756134e-02 2.182454e-14 2.201967e-03 3.286886e-02
4: 1.176240e-16 4.172401e-16 1.639750e-01 2.009148e-15 1.341245e-14
5: 3.764663e-02 4.555172e-02 2.208063e-14 9.908231e-04 3.416832e-02
---
1996: 1.450351e-16 7.944858e-16 8.565232e-05 2.933553e-15 1.928638e-14
1997: 2.288599e-16 6.115882e-16 2.543156e-05 3.089683e-15 1.835894e-14
1998: 1.723502e-06 4.795918e-14 6.936898e-03 6.338921e-02 1.623833e-11
1999: 1.763338e-16 6.891431e-16 1.045625e-05 3.424790e-15 1.718074e-14
2000: 1.788383e-02 3.520216e-08 2.683608e-14 3.496097e-03 2.714472e-07
K10 K11 K12 K13 K14
1: 2.161668e-01 1.640635e-02 7.412329e-15 1.103322e-02 7.621034e-03
2: 3.197928e-16 2.801745e-18 1.618601e-02 5.479059e-16 1.034362e-15
3: 1.112941e-02 2.341694e-02 4.522436e-15 6.236107e-02 6.998470e-03
4: 3.330870e-16 2.801745e-18 1.411016e-02 5.754238e-16 1.113395e-15
5: 6.136724e-03 1.718269e-02 4.662111e-15 9.602691e-02 1.615993e-03
---
1996: 7.586993e-16 2.801745e-18 2.646886e-02 9.283631e-16 1.872646e-15
1997: 3.772933e-16 2.801745e-18 1.184959e-05 8.639841e-16 1.535963e-15
1998: 4.408682e-14 7.028420e-14 2.506440e-03 1.803488e-07 2.313040e-12
1999: 4.582206e-16 2.801745e-18 1.385212e-09 1.236200e-15 1.361230e-15
2000: 1.150903e-05 7.643018e-14 8.742920e-15 9.467129e-03 4.035168e-05
broad_type symbol K01 K02 K03 K04
1: micro_immune PLXDC2 2.971594e-15 3.356741e-02 3.046753e-14 4.511489e-14
2: micro_immune DOCK4 2.756175e-15 4.358403e-03 3.044887e-14 4.541235e-14
3: micro_immune LRMDA 2.861045e-15 1.663837e-02 2.963549e-14 4.500699e-14
4: micro_immune FRMD4A 2.519641e-15 2.320878e-03 2.906520e-14 4.529767e-14
5: micro_immune ARHGAP24 2.917386e-15 5.605814e-03 3.049001e-14 4.529346e-14
---
1996: micro_immune WDR11 3.614304e-15 4.632794e-02 3.008912e-14 4.575960e-14
1997: micro_immune CXCR6 4.213801e-14 2.395118e-15 2.313236e-14 3.451279e-14
1998: micro_immune ANKRD13C 1.787968e-15 3.985755e-03 3.093736e-14 4.493011e-14
1999: micro_immune STS 4.762052e-14 6.450192e-16 3.201081e-02 3.647128e-14
2000: micro_immune MRPL16 4.499333e-14 3.719664e-16 2.535796e-02 3.805978e-14
K05 K06 K07 K08 K09
1: 2.787768e-14 4.536529e-02 6.927349e-02 6.645358e-02 2.576226e-14
2: 2.771203e-14 2.158446e-02 2.437016e-02 7.092336e-02 2.581333e-14
3: 2.703245e-14 1.598062e-02 5.751332e-02 9.652253e-02 2.465578e-14
4: 2.728917e-14 2.258228e-02 1.262044e-02 1.378379e-01 2.448238e-14
5: 2.746952e-14 1.227421e-02 8.875956e-02 5.140470e-02 2.558731e-14
---
1996: 3.323119e-14 9.464583e-03 3.246135e-03 4.987695e-05 2.801802e-14
1997: 3.565611e-02 1.310000e-15 1.215800e-15 1.859823e-15 4.252692e-14
1998: 3.076467e-14 1.577903e-04 4.375728e-10 2.123337e-03 2.784958e-14
1999: 3.611007e-14 3.243046e-16 3.660373e-16 2.910363e-16 7.802335e-05
2000: 1.319431e-02 1.092124e-16 1.185605e-16 2.518976e-16 3.101694e-13
K10 K11 K12
1: 4.284580e-03 3.681819e-14 1.356428e-02
2: 6.901694e-02 3.715605e-14 1.901650e-02
3: 4.032546e-03 3.599121e-14 8.674188e-03
4: 2.552147e-03 3.535350e-14 6.859777e-03
5: 1.118627e-02 3.741381e-14 1.494528e-02
---
1996: 3.260142e-03 3.873906e-14 5.072674e-14
1997: 1.518235e-15 4.017456e-14 1.375073e-15
1998: 4.038375e-02 3.426563e-14 5.132631e-14
1999: 2.856163e-16 3.520260e-14 3.546619e-18
2000: 3.810621e-16 4.208703e-03 1.209523e-17
broad_type symbol K01 K02 K03 K04
1: excitatory RORB 5.219785e-14 5.293619e-14 5.454697e-14 1.442586e-01
2: excitatory CUX2 5.131399e-14 5.392069e-14 6.178847e-02 5.056010e-14
3: excitatory IL1RAPL2 4.816518e-14 5.007263e-14 4.479094e-14 1.554495e-01
4: excitatory EPHA6 2.029934e-13 3.120833e-13 8.929167e-03 4.975029e-14
5: excitatory MEIS2 8.820663e-13 6.329072e-03 1.212507e-01 4.798831e-14
---
1996: excitatory CTNND1 8.744264e-08 6.687883e-09 6.168749e-08 2.941387e-02
1997: excitatory KLRD1 1.932750e-03 1.561749e-04 2.786574e-03 7.417891e-03
1998: excitatory ARCN1 6.058628e-08 6.412455e-06 7.208118e-05 1.343437e-03
1999: excitatory ACP2 5.137812e-14 1.778866e-09 1.772055e-06 2.038307e-10
2000: excitatory FAM81A 1.107749e-03 2.445363e-05 1.041774e-02 8.679680e-03
K05 K06 K07 K08 K09
1: 4.501555e-06 6.486706e-12 3.115896e-09 5.519004e-14 5.543619e-14
2: 6.092540e-14 1.036341e-01 3.165341e-11 5.306260e-14 5.716459e-14
3: 2.555155e-09 5.320712e-14 4.719281e-14 5.108116e-14 5.439786e-14
4: 6.600230e-14 1.915057e-01 4.080109e-11 5.373688e-14 5.525468e-14
5: 5.977909e-14 2.026135e-07 4.881992e-14 3.405423e-12 5.437775e-14
---
1996: 1.544422e-06 5.761799e-07 5.441063e-04 6.184462e-03 2.917239e-06
1997: 5.098564e-06 1.794280e-03 9.804848e-11 2.129459e-07 1.535254e-02
1998: 4.267390e-04 1.540929e-07 2.566357e-02 2.298687e-02 6.366950e-07
1999: 8.634757e-08 8.078235e-14 2.412781e-03 2.831961e-02 2.224490e-08
2000: 3.464352e-04 2.025131e-02 5.616256e-03 5.225664e-05 2.585959e-02
K10 K11 K12
1: 4.815794e-02 2.166414e-02 5.438591e-14
2: 1.152249e-08 7.476621e-06 5.310929e-14
3: 2.600455e-11 5.985460e-13 5.141438e-14
4: 2.889447e-10 1.277245e-11 5.524635e-14
5: 6.002455e-14 5.435667e-14 1.769529e-03
---
1996: 3.726710e-05 7.212366e-02 5.321210e-04
1997: 2.202306e-02 1.658209e-03 3.513505e-04
1998: 1.212861e-06 3.638522e-06 1.065393e-03
1999: 6.602095e-14 8.299553e-11 8.755509e-11
2000: 4.450144e-02 4.819948e-02 5.622873e-06
broad_type symbol K01 K02 K03 K04
1: inhibitory ADARB2 2.766723e-01 5.991360e-14 5.152359e-14 4.492247e-14
2: inhibitory CXCL14 1.379377e-01 5.571658e-14 4.773595e-14 4.462466e-14
3: inhibitory RELN 1.508838e-01 6.109794e-10 4.931309e-14 5.153243e-14
4: inhibitory GRIK3 3.130814e-14 6.348075e-14 4.667898e-14 4.686951e-14
5: inhibitory NFIB 1.413734e-01 7.127073e-14 8.140147e-04 5.046699e-14
---
1996: inhibitory AP3M2 8.741122e-04 1.222177e-04 3.274595e-03 5.849801e-02
1997: inhibitory GALNT7 5.033646e-05 2.794727e-06 1.058547e-01 5.321011e-05
1998: inhibitory PSMC4 2.801749e-10 7.015702e-14 1.469305e-07 7.831020e-03
1999: inhibitory PLEKHO1 5.596815e-09 1.960970e-10 1.029898e-04 1.548516e-02
2000: inhibitory RIMBP2 2.230634e-02 2.612701e-05 6.154704e-02 1.679517e-03
K05 K06 K07 K08 K09
1: 2.283793e-08 3.947332e-14 2.328855e-04 6.450310e-14 5.163735e-14
2: 5.604603e-14 3.343626e-14 2.259620e-10 5.872823e-14 5.196091e-14
3: 8.100962e-14 4.716569e-14 9.932720e-07 6.907554e-14 5.915468e-14
4: 4.915364e-14 4.229986e-02 3.983400e-14 2.987459e-10 5.255663e-14
5: 6.988242e-08 5.020048e-14 4.110753e-02 6.730828e-14 5.702409e-14
---
1996: 7.070438e-04 3.291708e-04 2.208931e-03 4.015153e-06 1.242075e-03
1997: 4.723046e-03 8.489098e-03 1.545351e-03 9.567540e-08 3.160690e-04
1998: 2.715698e-13 1.165791e-07 4.583328e-07 1.401585e-10 4.185940e-02
1999: 3.423962e-11 6.956701e-03 5.497471e-02 2.357856e-05 1.629559e-03
2000: 1.644017e-04 4.135851e-03 3.908925e-02 3.585112e-03 5.206685e-03
K10 K11 K12 K13 K14
1: 6.581096e-14 5.848218e-14 5.014993e-14 5.269369e-14 7.084830e-14
2: 5.748249e-14 5.548787e-14 4.617014e-14 4.848846e-14 6.675626e-14
3: 6.544875e-14 6.327432e-14 1.723515e-04 5.991200e-14 7.888176e-14
4: 6.218592e-14 1.420635e-12 7.535486e-02 5.671000e-14 6.399254e-14
5: 2.288153e-08 5.771135e-14 4.995881e-14 1.649013e-11 7.027149e-07
---
1996: 7.209483e-05 9.194262e-05 5.142262e-06 8.810467e-06 3.482595e-05
1997: 3.049385e-03 5.868211e-03 1.995241e-09 1.625130e-04 1.153223e-03
1998: 1.257456e-12 1.710540e-13 1.095095e-10 3.123679e-10 7.798892e-14
1999: 1.064204e-06 6.938298e-07 3.191425e-08 1.844853e-03 2.886933e-07
2000: 3.542726e-03 2.796509e-03 3.573793e-06 4.715719e-02 1.432758e-02
broad_type symbol K01 K02 K03 K04
1: astrocytes TNC 5.508544e-14 5.355202e-14 6.048987e-14 4.995452e-14
2: astrocytes CD44 4.826442e-07 4.937742e-14 5.950754e-14 3.523831e-12
3: astrocytes CABLES1 5.820693e-14 5.887997e-05 4.757135e-08 4.713368e-14
4: astrocytes WIF1 5.403599e-14 6.650400e-02 2.452031e-06 4.419095e-14
5: astrocytes CP 1.080412e-08 3.779148e-14 5.126544e-14 2.749986e-02
---
1996: astrocytes UBXN11 5.337963e-14 5.683441e-14 5.923437e-14 3.196347e-02
1997: astrocytes ATP6AP1L 4.828945e-06 2.902379e-02 5.262668e-03 1.926480e-02
1998: astrocytes PSMG4 2.861397e-07 3.263370e-05 5.375626e-06 2.400370e-03
1999: astrocytes AC010615.4 2.346774e-04 1.821599e-02 2.426731e-04 1.451961e-02
2000: astrocytes ADPRHL2 8.498947e-10 5.635027e-14 6.229237e-14 3.195369e-02
K05 K06 K07 K08 K09
1: 5.968234e-14 4.923187e-14 1.718578e-01 6.509417e-14 9.124448e-09
2: 6.024823e-14 4.912547e-14 7.937184e-02 6.758393e-14 6.462610e-14
3: 5.167050e-04 1.514491e-01 5.048680e-14 4.390321e-06 6.674808e-14
4: 6.562571e-14 3.256467e-02 4.461540e-14 2.496363e-11 6.560003e-14
5: 5.039962e-14 3.853433e-14 5.988266e-02 5.847123e-14 5.462482e-14
---
1996: 5.930296e-14 5.249242e-14 5.484194e-14 6.450261e-14 6.505828e-14
1997: 4.168829e-03 1.257576e-02 1.132977e-02 1.634542e-03 9.741721e-03
1998: 7.532285e-03 4.823328e-04 5.443867e-05 5.840001e-02 7.565350e-07
1999: 3.774251e-04 2.423429e-03 7.216716e-05 3.720462e-06 1.704744e-03
2000: 6.195603e-14 5.383538e-14 3.620362e-09 6.673996e-14 8.552366e-14
K10 K11 K12 K13 K14
1: 7.412629e-14 6.262882e-14 7.583952e-14 6.640169e-14 1.530919e-04
2: 7.141941e-14 6.491720e-14 5.565260e-14 6.617291e-14 1.095052e-01
3: 3.756261e-09 3.150442e-09 6.114988e-09 2.471356e-06 5.486905e-14
4: 7.839263e-14 3.319385e-12 5.992744e-14 2.627321e-07 4.706317e-14
5: 6.529651e-14 5.739577e-14 4.998159e-14 5.322422e-14 3.296692e-03
---
1996: 7.080094e-14 5.976464e-14 3.635713e-11 6.485558e-14 5.301219e-14
1997: 8.566273e-14 6.827795e-05 9.983806e-04 5.502123e-02 1.927157e-04
1998: 1.628670e-02 2.637286e-07 7.809776e-03 5.049444e-02 2.795895e-02
1999: 8.473280e-14 2.235145e-04 3.060273e-05 3.839901e-02 6.404636e-03
2000: 1.999261e-08 6.588056e-14 5.859846e-14 6.597820e-14 1.548130e-10
broad_type symbol K01 K02 K03
1: endo_stromal ST6GALNAC3 7.172008e-14 7.012454e-03 6.281933e-14
2: endo_stromal MECOM 7.203146e-14 5.680333e-02 6.299274e-14
3: endo_stromal SPARCL1 7.300515e-14 1.056339e-02 6.461209e-14
4: endo_stromal ELOVL7 7.136825e-14 5.803847e-03 6.229239e-14
5: endo_stromal DOCK9 7.182232e-14 4.991011e-03 6.308586e-14
---
1996: endo_stromal NOX4 5.978272e-14 6.474239e-14 1.374485e-12
1997: endo_stromal DPM1 7.257842e-14 4.533263e-04 6.354589e-14
1998: endo_stromal PPARGC1B 3.608014e-05 7.201470e-11 3.878628e-08
1999: endo_stromal PIM3 6.939683e-14 1.377866e-06 6.234442e-14
2000: endo_stromal DFFA 7.150331e-14 1.326031e-06 6.395182e-14
K04 K05 K06 K07 K08
1: 1.498302e-03 5.419028e-14 1.540184e-01 5.963086e-14 3.144429e-14
2: 1.236490e-02 5.492910e-14 5.139181e-02 6.014345e-14 3.331951e-14
3: 1.158405e-01 5.890062e-14 2.707504e-02 6.170423e-14 4.562174e-14
4: 4.569394e-04 5.283272e-14 1.364212e-01 5.894344e-14 2.882201e-14
5: 5.873066e-04 5.430258e-14 8.070333e-02 5.980567e-14 3.240603e-14
---
1996: 6.975139e-14 4.146344e-14 7.086873e-14 4.892767e-14 4.517662e-14
1997: 3.009232e-04 5.931691e-14 2.313132e-05 6.122265e-14 4.459234e-14
1998: 7.360141e-07 7.801095e-02 2.178976e-07 2.027907e-06 8.797911e-03
1999: 2.280931e-02 5.600714e-14 3.117069e-07 5.926342e-14 3.314743e-14
2000: 2.669230e-07 5.687601e-14 7.704663e-06 6.013716e-14 4.168505e-14
K09 K10 K11 K12 K13
1: 4.070986e-03 7.764734e-14 1.575869e-02 6.081887e-02 7.224379e-14
2: 3.891529e-03 7.805406e-14 1.164949e-02 5.016632e-02 7.239223e-14
3: 8.166619e-02 7.954355e-14 1.154343e-02 1.597070e-04 7.379952e-14
4: 1.002880e-03 7.678902e-14 1.164382e-02 2.856780e-02 7.164039e-14
5: 3.295122e-03 7.759341e-14 1.502902e-02 8.839820e-02 7.230092e-14
---
1996: 5.349982e-14 6.543058e-14 6.873331e-14 6.025480e-14 6.051628e-14
1997: 5.276834e-03 7.940677e-14 4.601825e-05 3.466727e-02 7.274670e-14
1998: 1.010242e-09 2.105622e-03 8.401897e-12 1.770163e-09 5.485598e-03
1999: 2.637981e-03 7.546770e-14 1.568118e-09 5.667143e-08 6.966712e-14
2000: 7.066721e-04 7.830004e-14 1.346238e-05 4.005797e-02 7.240697e-14
K14 K15 K16 K17 K18
1: 6.127805e-14 2.284951e-02 3.181706e-02 2.408074e-02 6.163245e-14
2: 6.152060e-14 6.260855e-02 2.273696e-02 2.504531e-02 6.328875e-14
3: 6.588025e-14 2.023795e-02 1.629141e-03 7.611883e-03 6.594707e-14
4: 5.991571e-14 1.159444e-02 8.649754e-02 2.305277e-02 6.077842e-14
5: 6.150957e-14 1.481647e-02 5.201353e-02 1.196794e-02 6.217186e-14
---
1996: 5.380636e-14 8.047601e-14 1.965383e-04 7.252267e-14 7.393960e-02
1997: 6.623165e-14 1.676482e-02 1.289766e-05 1.160637e-04 6.484637e-14
1998: 9.533727e-10 6.903673e-06 7.443941e-06 6.511035e-06 6.605045e-06
1999: 6.294554e-14 2.294094e-03 4.690200e-02 2.059725e-05 6.151744e-14
2000: 6.431186e-14 4.748313e-04 3.435668e-04 2.183126e-03 6.432454e-14
broad_type symbol K01 K02 K03 K04
1: microglia PRKY 6.770564e-14 2.503590e-05 5.318287e-14 8.432268e-12
2: microglia SERPINE1 3.279561e-04 5.538078e-14 3.039992e-07 5.671505e-03
3: microglia TMEM163 5.566815e-14 5.521313e-14 5.073022e-14 6.514299e-14
4: microglia P2RY12 2.028914e-12 1.539024e-01 4.951982e-14 2.482062e-04
5: microglia CX3CR1 5.511223e-14 1.305579e-01 4.749542e-14 8.788986e-05
---
1996: microglia UCK2 6.263724e-09 5.694415e-04 5.960537e-13 1.383746e-04
1997: microglia SPDYE1 5.642756e-14 5.520548e-03 5.403421e-05 1.160097e-05
1998: microglia HPS5 3.522207e-05 8.088349e-07 3.114857e-02 6.126449e-07
1999: microglia MEF2C 3.241175e-03 4.122203e-02 2.679070e-03 2.940959e-02
2000: microglia NHLH1 5.264484e-14 3.144937e-14 3.893406e-14 4.712589e-14
K05 K06 K07 K08 K09
1: 6.900231e-03 4.930286e-03 5.900456e-14 6.029417e-14 6.959494e-14
2: 5.823211e-14 7.216537e-02 6.057506e-14 5.697103e-14 6.911213e-14
3: 5.757283e-14 4.590347e-14 5.922927e-14 1.535616e-01 6.320724e-14
4: 6.169949e-14 5.040362e-14 6.216577e-14 3.801733e-08 6.861033e-14
5: 6.120999e-14 4.666223e-14 6.106205e-14 5.156635e-09 6.607159e-14
---
1996: 6.327394e-14 9.562713e-11 6.271645e-14 3.100203e-02 3.764879e-02
1997: 5.279486e-13 4.826186e-14 1.361846e-13 1.327453e-02 6.737341e-14
1998: 6.374449e-14 1.013532e-02 6.439315e-14 3.106419e-02 7.126954e-14
1999: 3.366089e-04 3.674704e-02 2.947039e-02 4.782318e-04 6.994018e-02
2000: 3.896336e-14 2.888198e-03 3.913413e-14 3.775779e-14 5.113412e-14
K10 K11 K12
1: 7.501088e-02 6.424984e-14 2.950255e-08
2: 7.063803e-14 6.239985e-14 1.996000e-13
3: 6.511028e-14 6.034444e-14 2.682775e-09
4: 9.049040e-07 6.477058e-14 6.606174e-08
5: 1.800022e-09 6.302235e-14 6.856641e-13
---
1996: 5.812981e-07 6.640436e-14 3.369117e-06
1997: 7.005570e-14 6.578345e-14 6.813124e-14
1998: 2.254832e-03 6.756021e-14 3.280228e-04
1999: 1.086983e-02 6.844830e-02 2.942337e-02
2000: 4.319230e-14 4.615849e-14 6.071050e-14
broad_type symbol K01 K02 K03 K04
1: immune THEMIS 1.033382e-06 1.308585e-01 1.575729e-12 8.768470e-03
2: immune CAMK4 2.996375e-12 9.535780e-02 4.159182e-14 7.775349e-02
3: immune MZB1 3.249409e-14 1.479838e-14 3.429147e-14 2.980185e-14
4: immune AOAH 1.324841e-07 1.351354e-01 4.565851e-14 1.174073e-04
5: immune PRKCH 1.600238e-06 1.274343e-01 4.366975e-12 6.585007e-04
---
1996: immune AC005833.1 3.432674e-14 2.340261e-14 3.533076e-14 3.208345e-14
1997: immune PBK 2.189431e-18 1.763067e-18 2.204274e-18 2.070107e-18
1998: immune ZNF266 5.631173e-02 1.617284e-03 4.686776e-14 3.946949e-14
1999: immune LHFPL5 4.771474e-14 1.552121e-03 1.347289e-05 4.004168e-14
2000: immune OGG1 6.451526e-02 8.146560e-11 4.247637e-14 3.959779e-14
K05 K06 K07 K08
1: 1.000951e-09 4.450650e-14 2.820396e-14 4.637007e-12
2: 2.224153e-10 4.318051e-14 2.484648e-14 4.259512e-14
3: 3.353328e-14 4.097989e-14 9.349851e-02 3.674409e-14
4: 2.842325e-04 4.818934e-07 3.326580e-14 2.180263e-05
5: 2.288542e-07 4.479990e-14 2.895130e-14 4.430732e-14
---
1996: 3.360024e-02 3.564273e-14 3.605409e-14 4.100123e-14
1997: 2.277688e-18 2.265090e-18 1.725560e-18 2.237138e-18
1998: 2.102843e-11 4.506053e-14 3.240238e-14 4.728729e-14
1999: 1.498920e-09 4.343182e-14 8.886839e-03 7.191751e-13
2000: 3.961926e-14 4.392731e-14 3.245002e-14 8.102254e-06devtools::session_info()Registered S3 method overwritten by 'cli':
method from
print.boxx spatstat.geom- Session info ---------------------------------------------------------------
setting value
version R version 4.0.5 (2021-03-31)
os CentOS Linux 7 (Core)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_US.UTF-8
ctype C
tz Europe/Zurich
date 2021-09-24
- Packages -------------------------------------------------------------------
! package * version date lib
abind 1.4-5 2016-07-21 [2]
assertthat * 0.2.1 2019-03-21 [2]
Biobase * 2.50.0 2020-10-27 [1]
BiocGenerics * 0.36.1 2021-04-16 [1]
BiocManager 1.30.16 2021-06-15 [1]
BiocParallel * 1.24.1 2020-11-06 [1]
BiocStyle * 2.18.1 2020-11-24 [1]
bitops 1.0-7 2021-04-24 [2]
bslib 0.3.0 2021-09-02 [2]
cachem 1.0.6 2021-08-19 [1]
Cairo 1.5-12.2 2020-07-07 [2]
callr 3.7.0 2021-04-20 [2]
circlize * 0.4.13 2021-06-09 [1]
cli 3.0.1 2021-07-17 [1]
clue 0.3-59 2021-04-16 [1]
cluster 2.1.2 2021-04-17 [2]
codetools 0.2-18 2020-11-04 [2]
colorout * 1.2-2 2021-04-15 [1]
colorspace 2.0-2 2021-06-24 [1]
ComplexHeatmap * 2.6.2 2020-11-12 [1]
cowplot 1.1.1 2020-12-30 [2]
crayon 1.4.1 2021-02-08 [2]
data.table * 1.14.0 2021-02-21 [2]
DBI 1.1.1 2021-01-15 [2]
DelayedArray 0.16.3 2021-03-24 [1]
deldir 0.2-10 2021-02-16 [2]
desc 1.3.0 2021-03-05 [2]
devtools 2.4.2 2021-06-07 [1]
digest 0.6.27 2020-10-24 [2]
dplyr 1.0.7 2021-06-18 [2]
ellipsis 0.3.2 2021-04-29 [2]
evaluate 0.14 2019-05-28 [2]
fansi 0.5.0 2021-05-25 [2]
fastmap 1.1.0 2021-01-25 [2]
fastmatch 1.1-3 2021-07-23 [1]
fgsea * 1.16.0 2020-10-27 [1]
fitdistrplus 1.1-5 2021-05-28 [2]
forcats * 0.5.1 2021-01-27 [2]
foreach 1.5.1 2020-10-15 [2]
fs 1.5.0 2020-07-31 [2]
future * 1.22.1 2021-08-25 [2]
future.apply 1.8.1 2021-08-10 [2]
generics 0.1.0 2020-10-31 [2]
GenomeInfoDb * 1.26.7 2021-04-08 [1]
GenomeInfoDbData 1.2.4 2021-04-15 [1]
GenomicRanges * 1.42.0 2020-10-27 [1]
GetoptLong 1.0.5 2020-12-15 [1]
ggplot.multistats * 1.0.0 2019-10-28 [1]
ggplot2 * 3.3.5 2021-06-25 [1]
ggrepel 0.9.1 2021-01-15 [2]
ggridges 0.5.3 2021-01-08 [2]
git2r 0.28.0 2021-01-10 [1]
GlobalOptions 0.1.2 2020-06-10 [1]
globals 0.14.0 2020-11-22 [2]
glue 1.4.2 2020-08-27 [2]
goftest 1.2-2 2019-12-02 [2]
gridExtra 2.3 2017-09-09 [2]
gtable 0.3.0 2019-03-25 [2]
here 1.0.1 2020-12-13 [2]
hexbin 1.28.2 2021-01-08 [2]
highr 0.9 2021-04-16 [2]
htmltools 0.5.2 2021-08-25 [2]
htmlwidgets 1.5.4 2021-09-08 [2]
httpuv 1.6.3 2021-09-09 [2]
httr 1.4.2 2020-07-20 [2]
ica 1.0-2 2018-05-24 [2]
igraph 1.2.6 2020-10-06 [2]
IRanges * 2.24.1 2020-12-12 [1]
irlba 2.3.3 2019-02-05 [2]
iterators 1.0.13 2020-10-15 [2]
jquerylib 0.1.4 2021-04-26 [2]
jsonlite 1.7.2 2020-12-09 [2]
KernSmooth 2.23-20 2021-05-03 [2]
knitr 1.34 2021-09-09 [1]
later 1.3.0 2021-08-18 [2]
lattice 0.20-44 2021-05-02 [2]
lazyeval 0.2.2 2019-03-15 [2]
R leiden 0.3.8 <NA> [2]
lifecycle 1.0.0 2021-02-15 [2]
listenv 0.8.0 2019-12-05 [2]
lmtest 0.9-38 2020-09-09 [2]
magick 2.7.2 2021-05-02 [2]
magrittr * 2.0.1 2020-11-17 [1]
MASS 7.3-54 2021-05-03 [2]
Matrix * 1.3-4 2021-06-01 [2]
MatrixGenerics * 1.2.1 2021-01-30 [1]
matrixStats * 0.60.1 2021-08-23 [1]
memoise 2.0.0 2021-01-26 [1]
mgcv 1.8-36 2021-06-01 [1]
mime 0.11 2021-06-23 [1]
miniUI 0.1.1.1 2018-05-18 [2]
munsell 0.5.0 2018-06-12 [2]
nlme 3.1-153 2021-09-07 [2]
parallelly 1.28.1 2021-09-09 [2]
patchwork 1.1.1 2020-12-17 [2]
pbapply 1.4-3 2020-08-18 [2]
pillar 1.6.2 2021-07-29 [1]
pkgbuild 1.2.0 2020-12-15 [1]
pkgconfig 2.0.3 2019-09-22 [2]
pkgload 1.2.2 2021-09-11 [2]
plotly 4.9.4.1 2021-06-18 [2]
plyr 1.8.6 2020-03-03 [2]
png 0.1-7 2013-12-03 [2]
polyclip 1.10-0 2019-03-14 [2]
prettyunits 1.1.1 2020-01-24 [2]
processx 3.5.2 2021-04-30 [2]
promises 1.2.0.1 2021-02-11 [2]
ps 1.6.0 2021-02-28 [2]
purrr * 0.3.4 2020-04-17 [2]
R6 2.5.1 2021-08-19 [2]
RANN 2.6.1 2019-01-08 [2]
rappdirs 0.3.3 2021-01-31 [2]
RColorBrewer * 1.1-2 2014-12-07 [2]
Rcpp 1.0.7 2021-07-07 [1]
RcppAnnoy 0.0.19 2021-07-30 [1]
RCurl 1.98-1.4 2021-08-17 [1]
registry 0.5-1 2019-03-05 [1]
remotes 2.4.0 2021-06-02 [1]
reshape2 1.4.4 2020-04-09 [2]
reticulate * 1.21 2021-09-14 [2]
rjson 0.2.20 2018-06-08 [1]
rlang 0.4.11 2021-04-30 [2]
rmarkdown 2.11 2021-09-14 [1]
ROCR 1.0-11 2020-05-02 [2]
rpart 4.1-15 2019-04-12 [2]
rprojroot 2.0.2 2020-11-15 [2]
Rtsne 0.15 2018-11-10 [2]
S4Vectors * 0.28.1 2020-12-09 [1]
sass 0.4.0 2021-05-12 [2]
scales * 1.1.1 2020-05-11 [2]
scattermore 0.7 2020-11-24 [2]
sctransform 0.3.2 2020-12-16 [2]
seriation * 1.3.0 2021-06-30 [1]
sessioninfo 1.1.1 2018-11-05 [1]
Seurat * 4.0.4 2021-08-20 [2]
SeuratObject * 4.0.2 2021-06-09 [2]
shape 1.4.6 2021-05-19 [1]
shiny 1.6.0 2021-01-25 [2]
SingleCellExperiment * 1.12.0 2020-10-27 [1]
spatstat.core 2.3-0 2021-07-16 [2]
spatstat.data 2.1-0 2021-03-21 [2]
spatstat.geom 2.2-2 2021-07-12 [2]
spatstat.sparse 2.0-0 2021-03-16 [2]
spatstat.utils 2.2-0 2021-06-14 [2]
stringi 1.7.4 2021-08-25 [1]
stringr * 1.4.0 2019-02-10 [2]
SummarizedExperiment * 1.20.0 2020-10-27 [1]
survival 3.2-13 2021-08-24 [2]
tensor 1.5 2012-05-05 [2]
testthat 3.0.4 2021-07-01 [2]
tibble 3.1.4 2021-08-25 [1]
tidyr 1.1.3 2021-03-03 [2]
tidyselect 1.1.1 2021-04-30 [2]
TSP 1.1-10 2020-04-17 [1]
usethis 2.0.1 2021-02-10 [1]
utf8 1.2.2 2021-07-24 [1]
uwot 0.1.10 2020-12-15 [2]
vctrs 0.3.8 2021-04-29 [2]
viridis * 0.6.1 2021-05-11 [1]
viridisLite * 0.4.0 2021-04-13 [1]
withr 2.4.2 2021-04-18 [2]
workflowr * 1.6.2 2020-04-30 [1]
writexl * 1.4.0 2021-04-20 [1]
xfun 0.26 2021-09-14 [1]
xtable 1.8-4 2019-04-21 [2]
XVector 0.30.0 2020-10-27 [1]
yaml 2.2.1 2020-02-01 [2]
zlibbioc 1.36.0 2020-10-27 [1]
zoo 1.8-9 2021-03-09 [2]
source
CRAN (R 4.0.0)
CRAN (R 4.0.0)
Bioconductor
Bioconductor
CRAN (R 4.0.3)
Bioconductor
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Github (jalvesaq/colorout@79931fd)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.2)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
Bioconductor
Bioconductor
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.2)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
<NA>
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.5)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.1)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.5)
CRAN (R 4.0.0)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.1)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
[1] /pstore/home/macnairw/lib/conda_r3.12
[2] /pstore/home/macnairw/.conda/envs/r_4.0.3/lib/R/library
R -- Package was removed from disk.
sessionInfo()R version 4.0.5 (2021-03-31)
Platform: x86_64-conda-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /pstore/home/macnairw/.conda/envs/r_4.0.3/lib/libopenblasp-r0.3.12.so
locale:
[1] LC_CTYPE=C LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] grid parallel stats4 stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] writexl_1.4.0 reticulate_1.21
[3] fgsea_1.16.0 BiocParallel_1.24.1
[5] ggplot.multistats_1.0.0 seriation_1.3.0
[7] ComplexHeatmap_2.6.2 SeuratObject_4.0.2
[9] Seurat_4.0.4 future_1.22.1
[11] Matrix_1.3-4 SingleCellExperiment_1.12.0
[13] SummarizedExperiment_1.20.0 Biobase_2.50.0
[15] GenomicRanges_1.42.0 GenomeInfoDb_1.26.7
[17] IRanges_2.24.1 S4Vectors_0.28.1
[19] BiocGenerics_0.36.1 MatrixGenerics_1.2.1
[21] matrixStats_0.60.1 purrr_0.3.4
[23] forcats_0.5.1 ggplot2_3.3.5
[25] scales_1.1.1 viridis_0.6.1
[27] viridisLite_0.4.0 assertthat_0.2.1
[29] stringr_1.4.0 data.table_1.14.0
[31] magrittr_2.0.1 circlize_0.4.13
[33] RColorBrewer_1.1-2 BiocStyle_2.18.1
[35] colorout_1.2-2 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] utf8_1.2.2 tidyselect_1.1.1 htmlwidgets_1.5.4
[4] TSP_1.1-10 Rtsne_0.15 devtools_2.4.2
[7] munsell_0.5.0 codetools_0.2-18 ica_1.0-2
[10] miniUI_0.1.1.1 withr_2.4.2 colorspace_2.0-2
[13] highr_0.9 knitr_1.34 ROCR_1.0-11
[16] tensor_1.5 listenv_0.8.0 git2r_0.28.0
[19] GenomeInfoDbData_1.2.4 polyclip_1.10-0 rprojroot_2.0.2
[22] parallelly_1.28.1 vctrs_0.3.8 generics_0.1.0
[25] xfun_0.26 R6_2.5.1 clue_0.3-59
[28] bitops_1.0-7 spatstat.utils_2.2-0 cachem_1.0.6
[31] DelayedArray_0.16.3 promises_1.2.0.1 gtable_0.3.0
[34] Cairo_1.5-12.2 globals_0.14.0 processx_3.5.2
[37] goftest_1.2-2 rlang_0.4.11 GlobalOptions_0.1.2
[40] splines_4.0.5 lazyeval_0.2.2 hexbin_1.28.2
[43] spatstat.geom_2.2-2 BiocManager_1.30.16 yaml_2.2.1
[46] reshape2_1.4.4 abind_1.4-5 httpuv_1.6.3
[49] usethis_2.0.1 tools_4.0.5 ellipsis_0.3.2
[52] spatstat.core_2.3-0 jquerylib_0.1.4 sessioninfo_1.1.1
[55] ggridges_0.5.3 Rcpp_1.0.7 plyr_1.8.6
[58] zlibbioc_1.36.0 RCurl_1.98-1.4 prettyunits_1.1.1
[61] ps_1.6.0 rpart_4.1-15 deldir_0.2-10
[64] pbapply_1.4-3 GetoptLong_1.0.5 cowplot_1.1.1
[67] zoo_1.8-9 ggrepel_0.9.1 cluster_2.1.2
[70] fs_1.5.0 here_1.0.1 magick_2.7.2
[73] scattermore_0.7 lmtest_0.9-38 RANN_2.6.1
[76] fitdistrplus_1.1-5 pkgload_1.2.2 patchwork_1.1.1
[79] mime_0.11 evaluate_0.14 xtable_1.8-4
[82] gridExtra_2.3 shape_1.4.6 testthat_3.0.4
[85] compiler_4.0.5 tibble_3.1.4 KernSmooth_2.23-20
[88] crayon_1.4.1 htmltools_0.5.2 mgcv_1.8-36
[91] later_1.3.0 tidyr_1.1.3 DBI_1.1.1
[94] MASS_7.3-54 rappdirs_0.3.3 cli_3.0.1
[97] igraph_1.2.6 pkgconfig_2.0.3 registry_0.5-1
[100] plotly_4.9.4.1 spatstat.sparse_2.0-0 foreach_1.5.1
[103] bslib_0.3.0 XVector_0.30.0 callr_3.7.0
[106] digest_0.6.27 sctransform_0.3.2 RcppAnnoy_0.0.19
[109] spatstat.data_2.1-0 rmarkdown_2.11 leiden_0.3.8
[112] fastmatch_1.1-3 uwot_0.1.10 shiny_1.6.0
[115] rjson_0.2.20 lifecycle_1.0.0 nlme_3.1-153
[118] jsonlite_1.7.2 desc_1.3.0 fansi_0.5.0
[121] pillar_1.6.2 lattice_0.20-44 fastmap_1.1.0
[124] httr_1.4.2 pkgbuild_1.2.0 survival_3.2-13
[127] remotes_2.4.0 glue_1.4.2 png_0.1-7
[130] iterators_1.0.13 stringi_1.7.4 sass_0.4.0
[133] memoise_2.0.0 dplyr_1.0.7 irlba_2.3.3
[136] future.apply_1.8.1