Checks: Oligo-PC1 muscat run
Will Macnair
Institute for Molecular Life Sciences, University of Zurich, SwitzerlandSwiss Institute of Bioinformatics (SIB), University of Zurich, SwitzerlandJuly 02, 2021
Last updated: 2021-07-02
Checks: 4 3
Knit directory: MS_lesions/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
The global environment had objects present when the code in the R Markdown file was run. These objects can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment. Use wflow_publish or wflow_build to ensure that the code is always run in an empty environment.
The following objects were defined in the global environment when these results were created:
| Name | Class | Size |
|---|---|---|
| q | function | 1008 bytes |
The command set.seed(20210118) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
- calc_oligo_pcs
- calc_patient_level_gsea
- calc_patient_level_muscat
- calc_pseudo_bulk
- calc_soup
- calc_tweaked_inputs
- load_outputs_for_plotting
- load_standard_things
- plot_de_barplot
- plot_gsea_results_test
- plot_olg_pcs
- plot_olg_pcs_annot
- plot_olg_pcs_vs_qc_stats
- plot_oligo_pc_heatmap
- plot_oligo_pc_scatters_down
- plot_oligo_pc_scatters_up
- plot_soup_vs_results
- plot_top_genes_oligo_pc_common
- plot_top_genes_oligo_pc_specific
- save_outputs
- session_info
- session-info-chunk-inserted-by-workflowr
- setup_input
- setup_outputs
To ensure reproducibility of the results, delete the cache directory supp10_muscat_olg_pc1_cache and re-run the analysis. To have workflowr automatically delete the cache directory prior to building the file, set delete_cache = TRUE when running wflow_build() or wflow_publish().
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 0ec42e1. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rprofile
Ignored: .Rproj.user/
Ignored: ._.DS_Store
Ignored: ._MS_lesions.sublime-project
Ignored: .log/
Ignored: MS_lesions.sublime-project
Ignored: MS_lesions.sublime-workspace
Ignored: analysis/.__site.yml
Ignored: analysis/ms02_doublet_id_cache/
Ignored: analysis/ms03_SampleQC_cache/
Ignored: analysis/ms04_conos_cache/
Ignored: analysis/ms05_splitting_cache/
Ignored: analysis/ms06_sccaf_cache/
Ignored: analysis/ms07_soup_cache/
Ignored: analysis/ms08_modules_cache/
Ignored: analysis/ms09_ancombc_cache/
Ignored: analysis/ms09_ancombc_clean_1e3_cache/
Ignored: analysis/ms09_ancombc_clean_2e3_cache/
Ignored: analysis/ms10_muscat_run01_cache/
Ignored: analysis/ms10_muscat_run02_cache/
Ignored: analysis/ms10_muscat_template_broad_cache/
Ignored: analysis/ms10_muscat_template_fine_cache/
Ignored: analysis/ms11_paga_cache/
Ignored: analysis/ms12_markers_cache/
Ignored: analysis/ms13_labelling_cache/
Ignored: analysis/ms14_lesions_cache/
Ignored: analysis/supp06_sccaf_cache/
Ignored: analysis/supp09_ancombc_cache/
Ignored: analysis/supp10_muscat_cache/
Ignored: analysis/supp10_muscat_heatmaps_cache/
Ignored: analysis/supp10_muscat_olg_pc1_cache/
Ignored: analysis/supp10_muscat_olg_pc2_cache/
Ignored: analysis/supp10_muscat_olg_pc_cache/
Ignored: code/muscat_plan.txt
Ignored: data/
Ignored: figures/
Ignored: output/
Untracked files:
Untracked: Rplots.pdf
Untracked: analysis/supp10_muscat_heatmaps.Rmd
Untracked: analysis/supp10_muscat_olg_pc1.Rmd
Untracked: analysis/supp10_muscat_olg_pc2.Rmd
Untracked: analysis/temp_igll5.R
Untracked: code/dev_muscat_fixing_zero_conditions_2021-06-28.R
Untracked: code/dev_stan_cauchy_fits_2021-06-28.R
Untracked: code/supp10_muscat_olg_pc.R
Unstaged changes:
Modified: analysis/index.Rmd
Modified: analysis/ms10_muscat_template_broad.Rmd
Modified: analysis/ms10_muscat_template_fine.Rmd
Modified: analysis/ms12_markers.Rmd
Modified: analysis/supp09_ancombc.Rmd
Modified: code/ms07_soup.R
Modified: code/ms09_ancombc.R
Modified: code/ms10_muscat_fns.R
Modified: code/ms10_muscat_runs.R
Modified: code/ms12_markers.R
Deleted: code/supp08_ancombc.R
Modified: code/supp10_muscat.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
Setup / definitions
Libraries
Helper functions
source('code/ms00_utils.R')
source('code/ms03_SampleQC.R')
source('code/ms07_soup.R')
source('code/ms09_ancombc.R')
source('code/ms10_muscat_runs.R')
source('code/supp10_muscat_olg_pc.R')Inputs
# define inputs
sce_f = 'data/sce_raw/ms_sce.rds'
meta_f = 'data/metadata/metadata_updated_20201127.txt'
labels_f = 'data/byhand_markers/validation_markers_2021-05-31.csv'
labelled_f = 'output/ms13_labelling/conos_labelled_2021-05-31.txt.gz'
# get soup values
soup_dir = 'output/ms07_soup'
pb_soup_f = file.path(soup_dir, 'pb_soup_broad_maximum_2021-06-01.rds')
pb_orig_f = file.path(soup_dir, 'pb_sum_broad_2021-06-01.rds')
# get qc values
qc_dir = 'output/ms03_SampleQC'
qc_f = file.path(qc_dir, 'ms_qc_dt.txt')Outputs
# ancombc variables
sample_vars = c('sample_id', 'matter', 'lesion_type',
'neuro_ok', 'neuro_prop', 'source', 'patient_id',
'sex', 'age_norm', 'pmi_cat')
mad_cut = 2
# define variables and celltypes for excluding oligo_D etc
d_vars = c("source", "patient_id", "sex", "age_norm")
d_types = c('Oligo_D', 'COP_A2')
# define pseudobulk files
pb_olg_pc_f = sprintf('%s/pb_olg_pc_sum_broad_%s.rds', soup_dir, '2021-06-30')
# define muscat runs
date_tag = '2021-07-01'
cluster_var = 'type_broad'
method_spec = list(method = 'edgeR', treat = FALSE)
filter = 'both'
padj_cut = 0.05
fc_cut = 0
fgsea_cut = 0.05
min_cells = 10
soup_cut = 0.1
n_cores = 8
subset_d = list(matter = 'WM', neuro_ok = TRUE, oligo_d = FALSE)
# outputs directory
save_dir = 'output/ms10_muscat/run14'
if (!dir.exists(save_dir))
dir.create(save_dir)
# define files to save
formula_str = '~ oligo_pc1 + sex + age_norm + pmi_cat'
run_tag = 'olg_pc1'
this_var = 'oligo_pc1'
olgpc_res_f = sprintf('%s/muscat_res_dt_%s_%s.txt.gz', save_dir, run_tag, date_tag)
fgsea_pat = sprintf('%s/fgsea_dt_%s_%s_%s.txt.gz', save_dir, run_tag, '%s', '%s')
fgsea_fs = sapply(names(paths_list),
function(n) sprintf(fgsea_pat, n, date_tag))Load inputs
labels_dt = load_names_dt(labels_f) %>%
.[, cluster_id := type_fine]
meta_dt = load_meta_dt(meta_f)
conos_dt = load_labelled_dt(labelled_f, labels_f) %>%
merge(meta_dt, by = 'sample_id') %>%
add_neuro_props(mad_cut = mad_cut)
qc_stats = calc_qc_stats(qc_dir, qc_f,
conos_dt[, .(cell_id, sample_id, type_broad, type_fine, conos)]) %>%
merge(meta_dt[, .(sample_id, lesion_type, patient_id)], by = 'sample_id')Processing / calculations
props_wm_all = calc_props_wm_all(conos_dt, sample_vars)
olg_pc_dt = calc_olg_pc_dt(conos_dt, d_types, props_wm_all)pat_annots = calc_patient_annots(conos_dt)
conos_tweak = calc_conos_tweak(labelled_f, labels_f, meta_dt,
mad_cut, d_types)
meta_tweak = calc_meta_tweak(meta_f, olg_pc_dt, pat_annots)pb_olg_pc = make_pb_object(pb_olg_pc_f, sce_f, meta_tweak, conos_tweak,
cluster_var = cluster_var, fun = 'sum', n_cores = n_cores)loading pre-saved objectcols_dt = pb_olg_pc %>% colData %>% as.data.frame %>%
as.data.table(keep.rownames = 'sample_id')pb_soup = pb_soup_f %>% readRDS
pb_orig = pb_orig_f %>% readRDS %>% .subset_pb(list(matter = c('WM'))) subsetting pb object restricting to samples that meet subset criteria updating factors to remove levels no longer observedcontam_dt = .calc_contam_dt(pb_soup, pb_orig, min_cells)res_dt = calc_patient_muscat(olgpc_res_f, pb_olg_pc, contam_dt,
gtf_f, formula_str, method_spec, filter, min_cells,
padj_cut, fc_cut, soup_cut, n_cores)calc_patient_level_gsea(olgpc_res_f, fgsea_pat, fgsea_cut, date_tag, n_cores)already done!NULLlabels_dt = .load_labels_dt(labels_f, cluster_var)
magma_dt = .load_magma_dt(magma_f, pb_orig)
tfs_dt = .load_tfs_dt(tfs_f, pb_orig)
lof_dt = .load_lof_dt(lof_f, pb_orig)
coloc_dt = .load_coloc_dt(coloc_f, pb_orig)
# get muscat results
res_all = olgpc_res_f %>% fread
res_all[, logCPM := logCPM - min(min(logCPM), 0), by = cluster_id]
res_dt = .load_muscat_results(res_all, labels_dt, params)
# get fgsea results
fgsea_list = .load_fgseas_list(fgsea_fs, labels_dt)
# prep for stacked bars
signif_dt = res_dt[ updown_soup != 'insignif' & !is.na(p_adj.soup) ]
assert_that(all(abs(signif_dt$logFC) >= fc_cut))[1] TRUEuniques_dt = .calc_uniques_dt(signif_dt)
stacked_dt = .calc_stacked_dt(uniques_dt)
# restrict to significant genes only
message(' getting top genes') getting top genestop_dt = .calc_top_genes(signif_dt, res_dt, uniques_dt,
magma_dt, tfs_dt, lof_dt, coloc_dt, fc_cut, top_n = 30)
top_gwas_dt = .calc_top_gwas_dt(res_dt, uniques_dt, magma_dt,
tfs_dt, lof_dt, coloc_dt, fc_cut = NULL, magma_cut = 0.05)
# uniques_dt[ grepl('Imm', cluster_id) & shared_outside_cl == FALSE ] %>%
# merge(res_dt, by = c('cluster_id', 'gene_id', 'symbol', 'var_type', 'test_var')) %>%
# .[, .(symbol, logFC = round(logFC, 2), p_adj.soup)] %>% .[ order(logFC) ]
# fgsea_dt[ grepl('PLOD1', leadingEdge) & coef == 'oligo_pc1' & padj < 0.5,
# .(cluster_id, pathway = str_match(pathway, gsea_regex)[, 3] %>% tolower,
# main_path, padj, NES) ] %>%
# .[order(cluster_id, padj)] %>% print(200)CLR plots
CLRs to show Oligo_D outliers
input_all = conos_dt[(matter == "WM") &
(type_broad %in% c('Oligodendrocytes', 'OPCs / COPs'))]
cat('### All\n')All
(plot_patient_clrs(input_all, props_wm_all))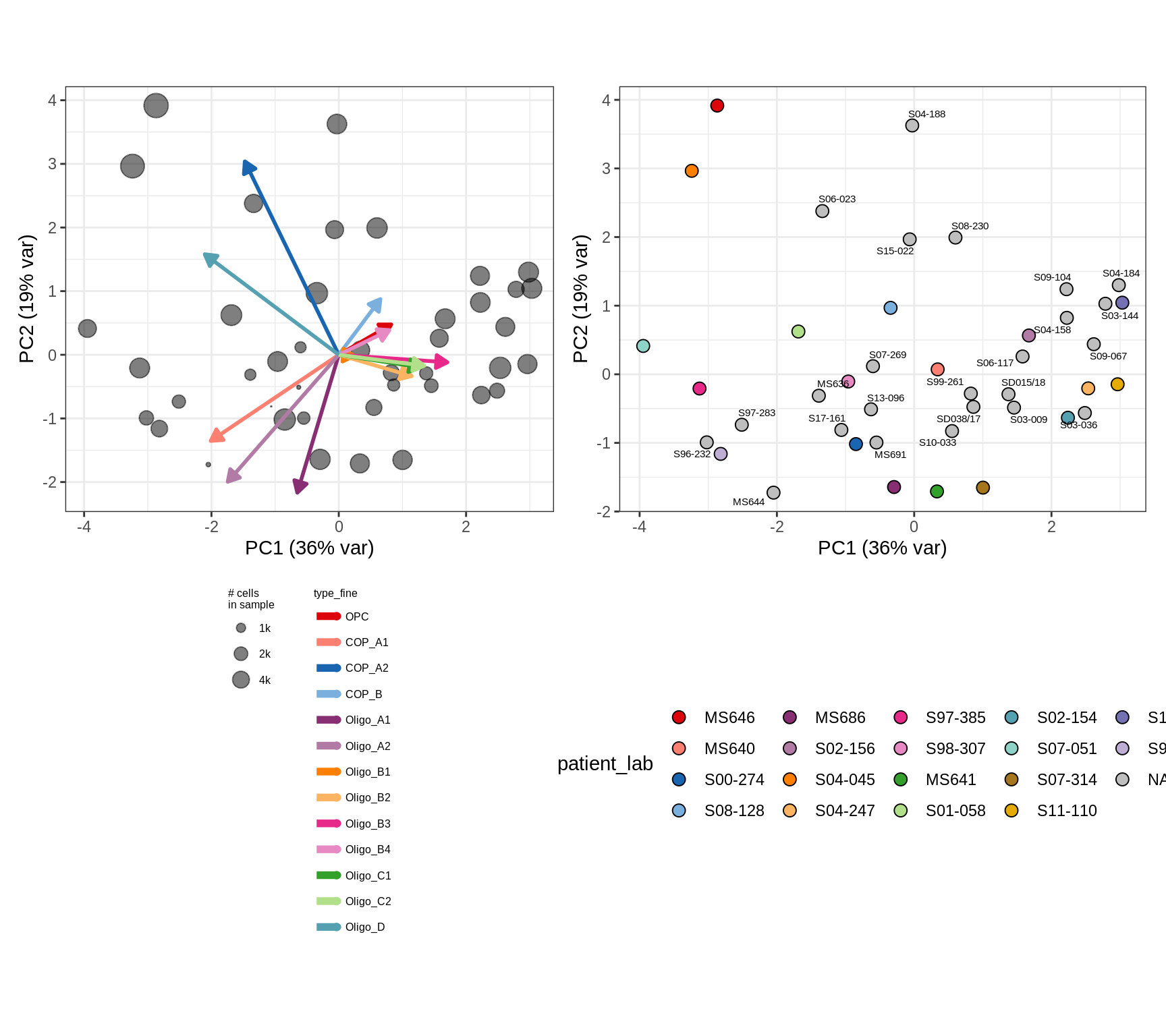
cat('\n\n')cat('### Oligo_D + COP_A2 excluded\n')Oligo_D + COP_A2 excluded
input_sel = input_all[ !(type_fine %in% d_types) ]
(plot_patient_clrs(input_sel, props_wm_all))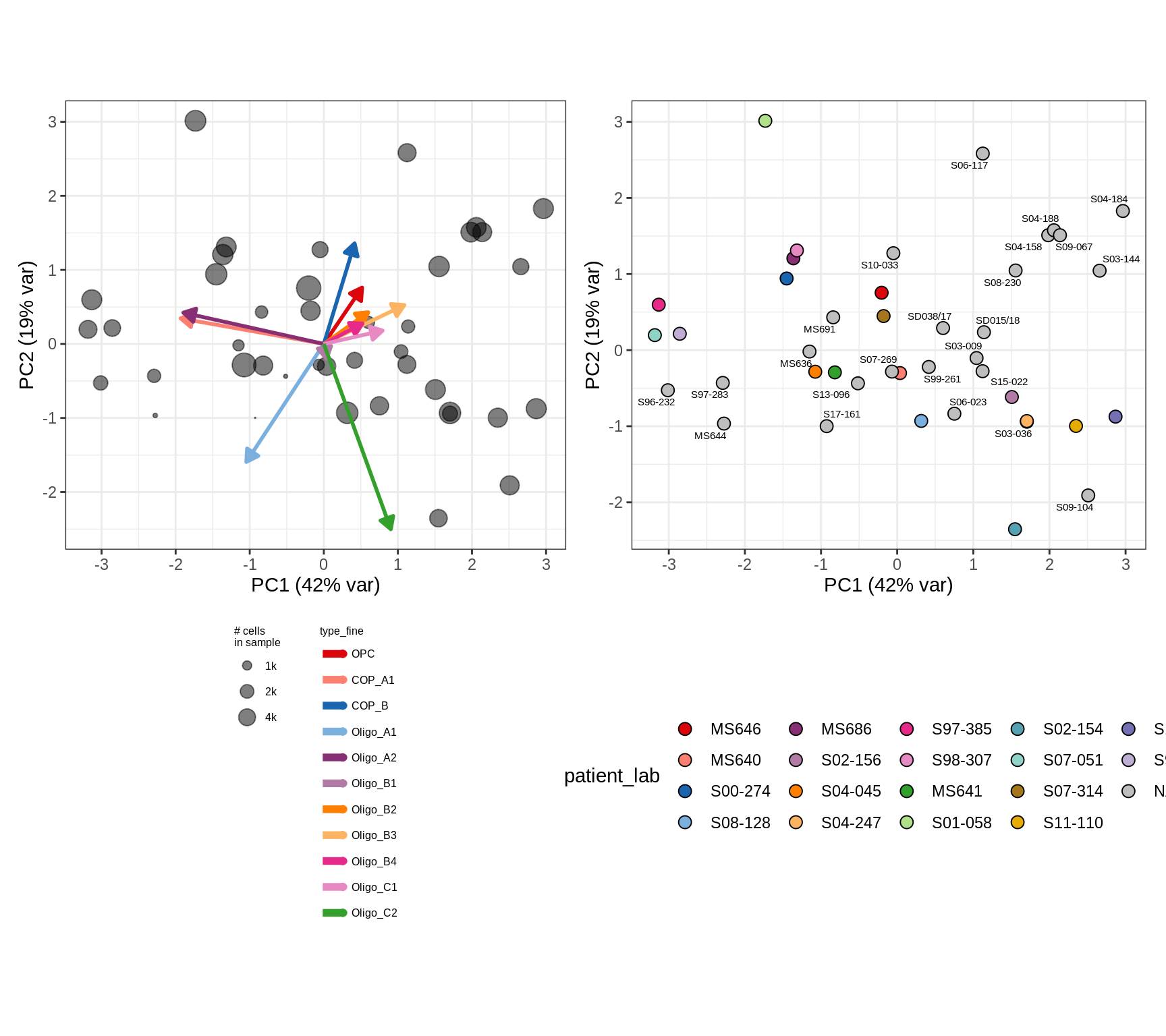
cat('\n\n')Same CLRs, annotated
cat('### All\n')All
(plot_patient_clrs_annots(input_all, props_wm_all, meta_tweak))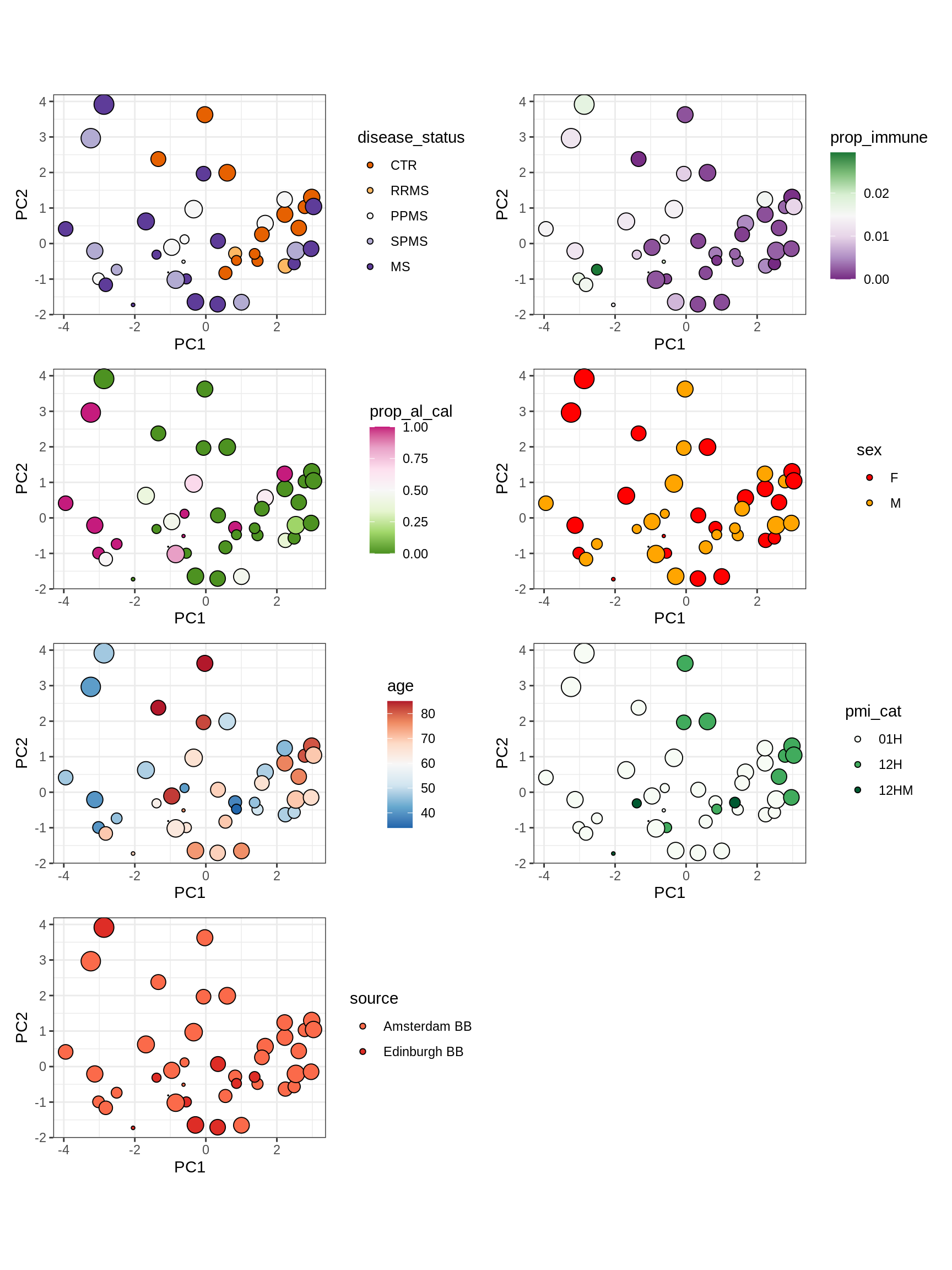
cat('\n\n')cat('### Oligo_D + COP_A2 excluded\n')Oligo_D + COP_A2 excluded
input_sel = input_all[ !(type_fine %in% d_types) ]
(plot_patient_clrs_annots(input_sel, props_wm_all, meta_tweak))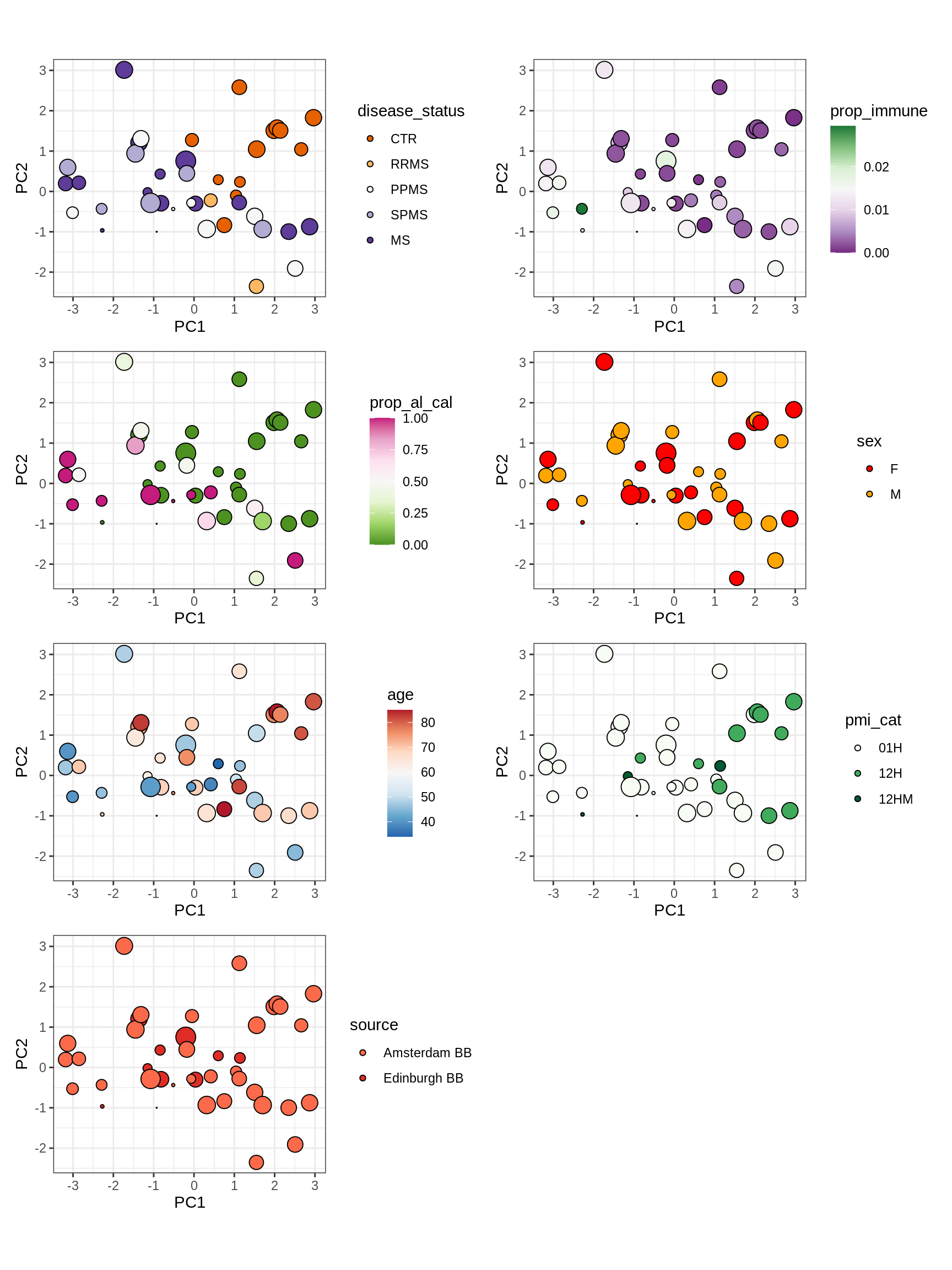
cat('\n\n')Muscat summary
Barplot of significant genes by contrast
tmp_dt = copy(stacked_dt)
print(plot_n_signif_barplot(tmp_dt, facet_by = 'cluster_id'))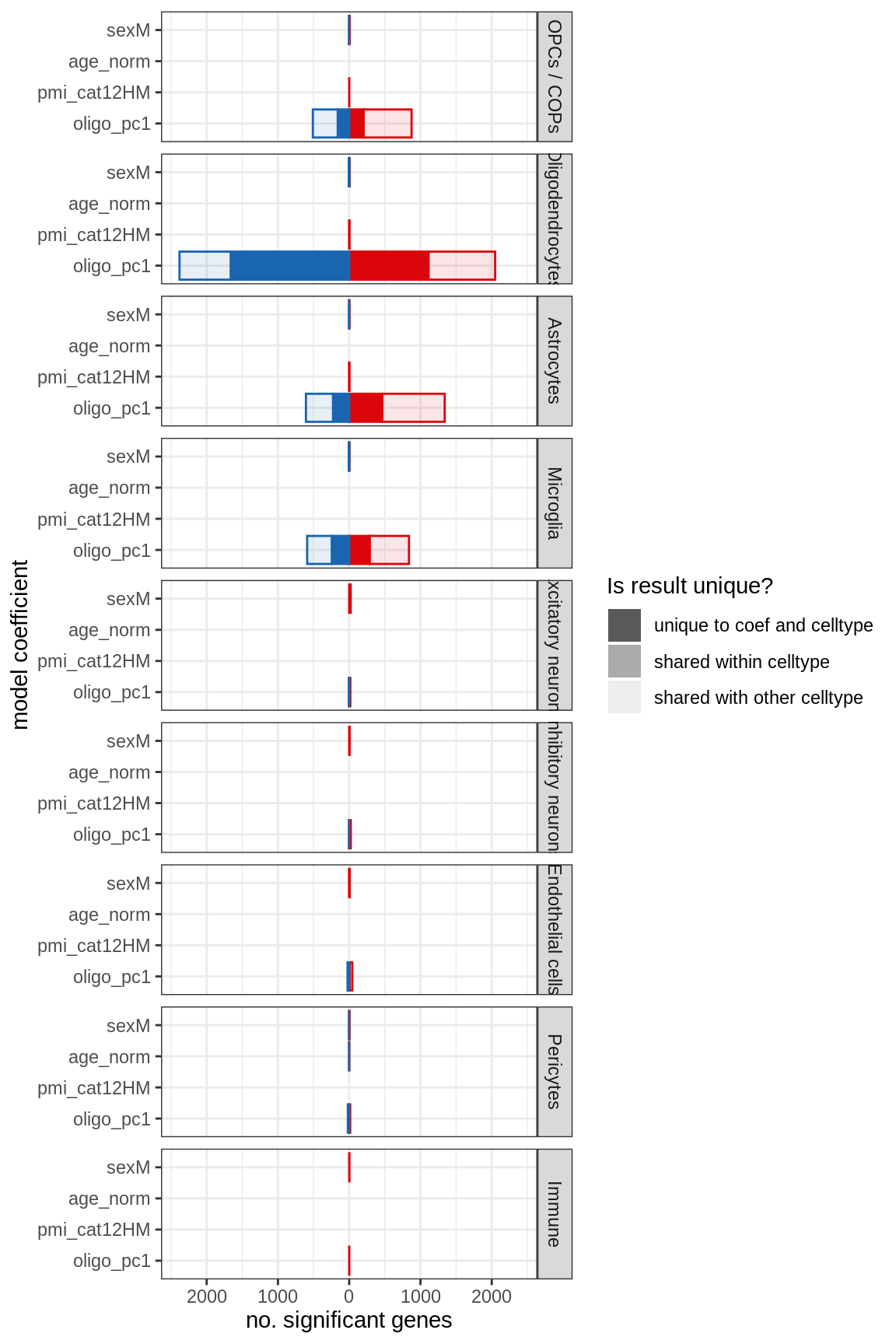
this_type = 'test'
how_unique = 'celltype_specific'
cat('# Top ', how_unique, ' genes, ', this_type,
' variables{.tabset}\n', sep='')Top celltype_specific genes, test variables
for (cl in levels(top_dt$cluster_id)) {
tmp_dt = top_dt[ unique_var %in% unique_ls[[how_unique]] &
cluster_id == cl & test_var == this_var ]
tb = unique(tmp_dt$type_broad)
if ( nrow(tmp_dt) == 0 )
next
cat('## ', cl, '\n')
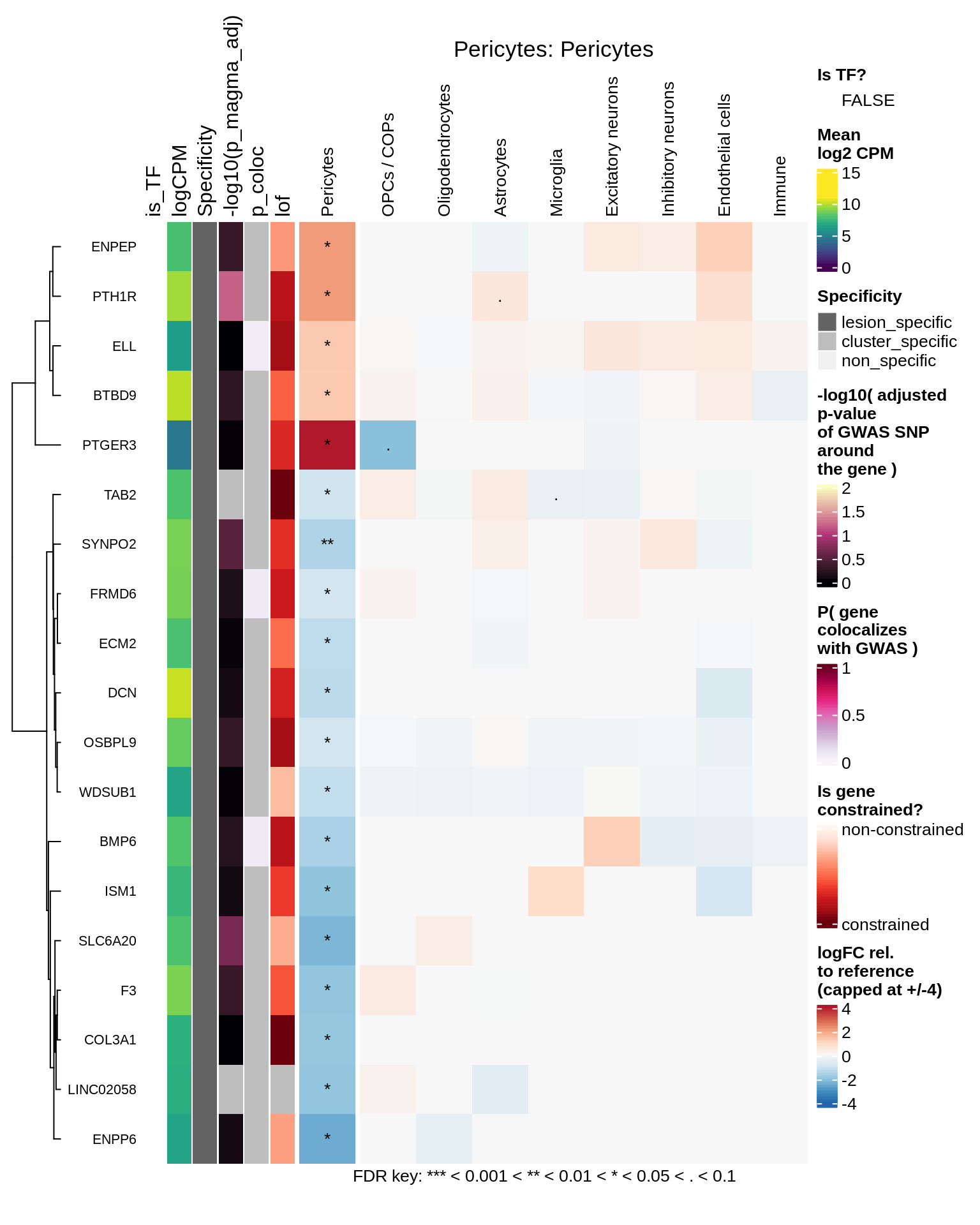
hm_obj = plot_top_genes_heatmap_fn_across_celltypes(tmp_dt, res_dt, labels_dt,
max_fc = 4, title = paste(tb, cl, sep=': '))
draw(hm_obj, padding = unit(c(0.5, 0.1, 0.1, 0.1), "in"))
.add_fdr_legend('logfc')
cat('\n\n')
}OPCs / COPs
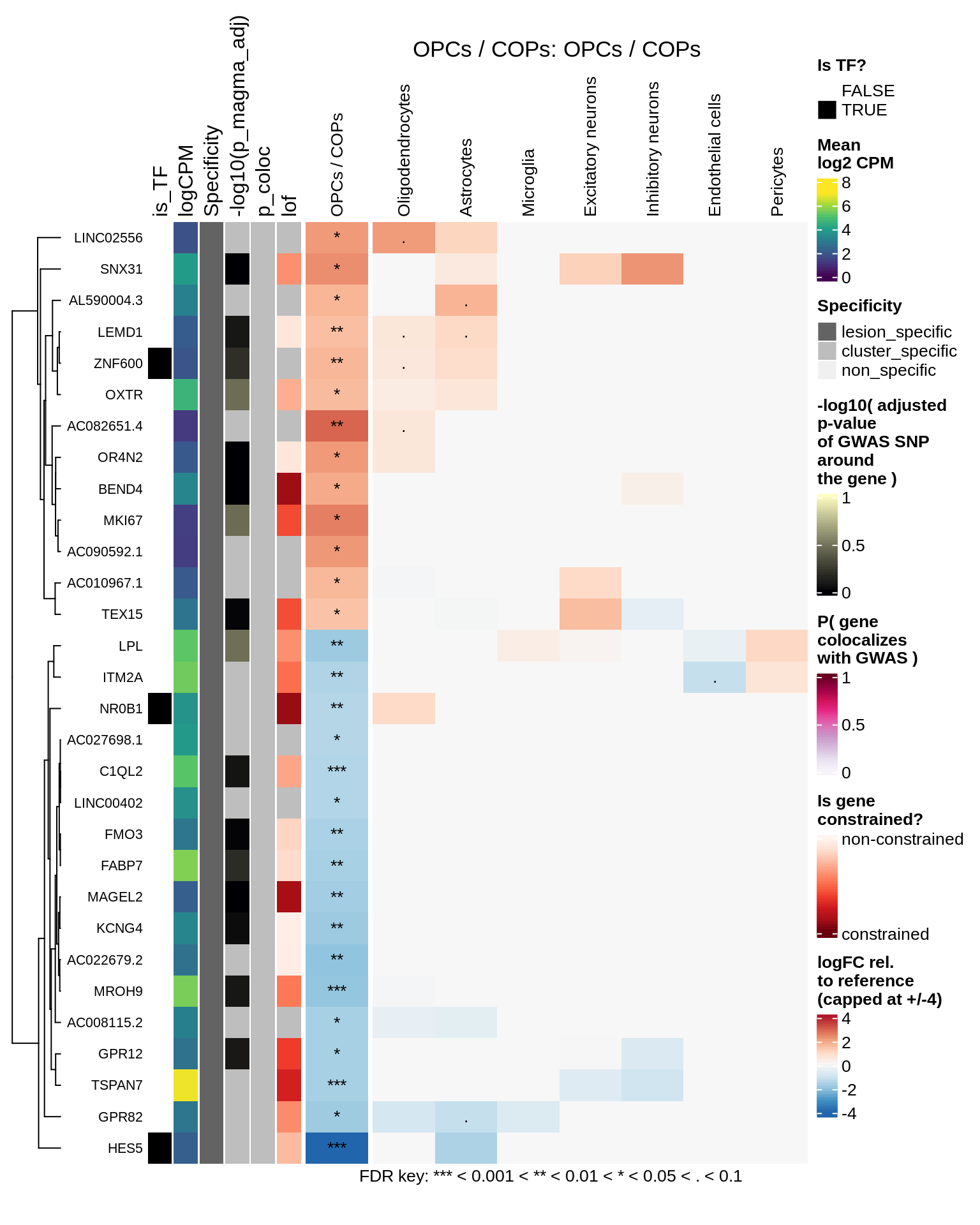
Oligodendrocytes
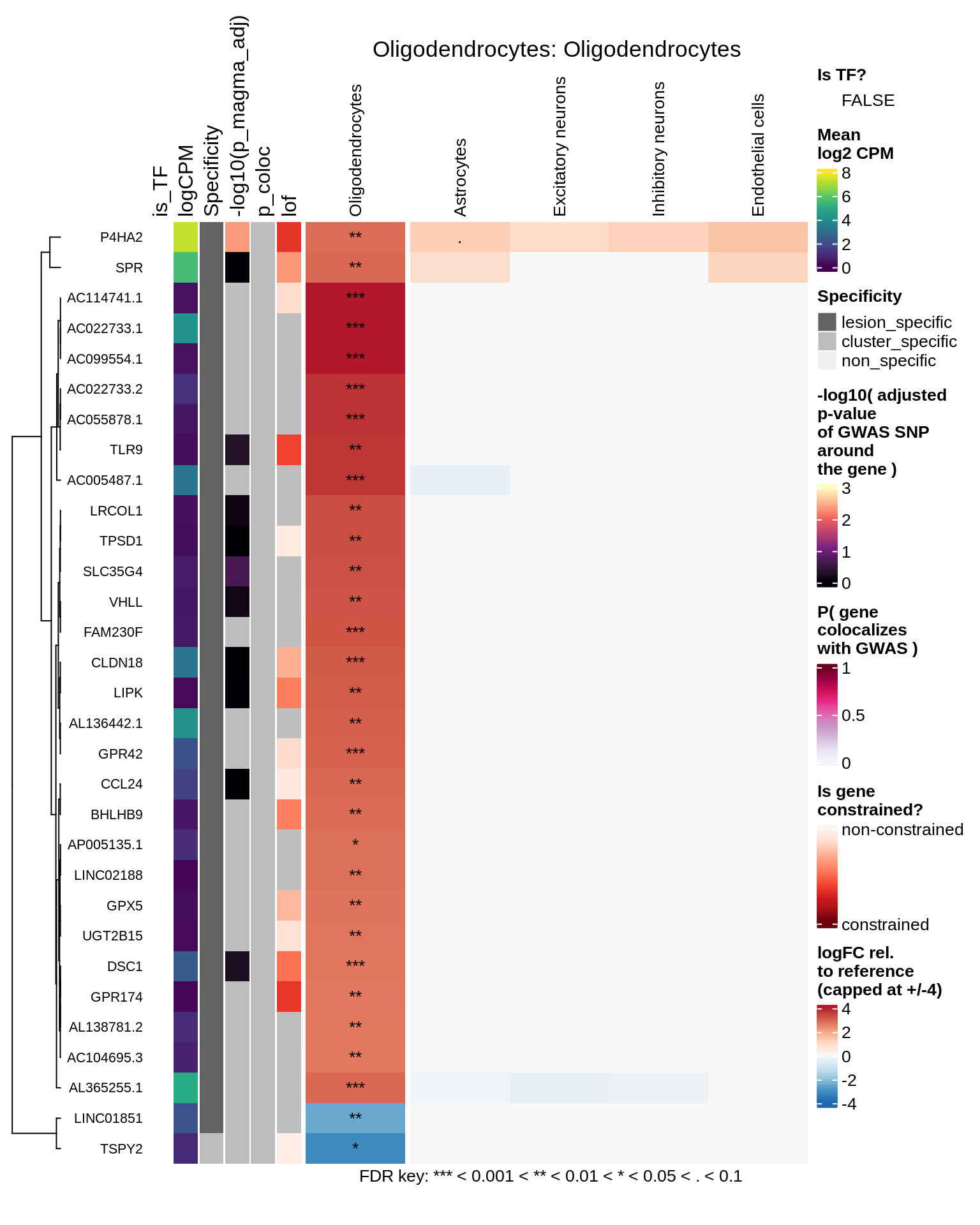
Astrocytes
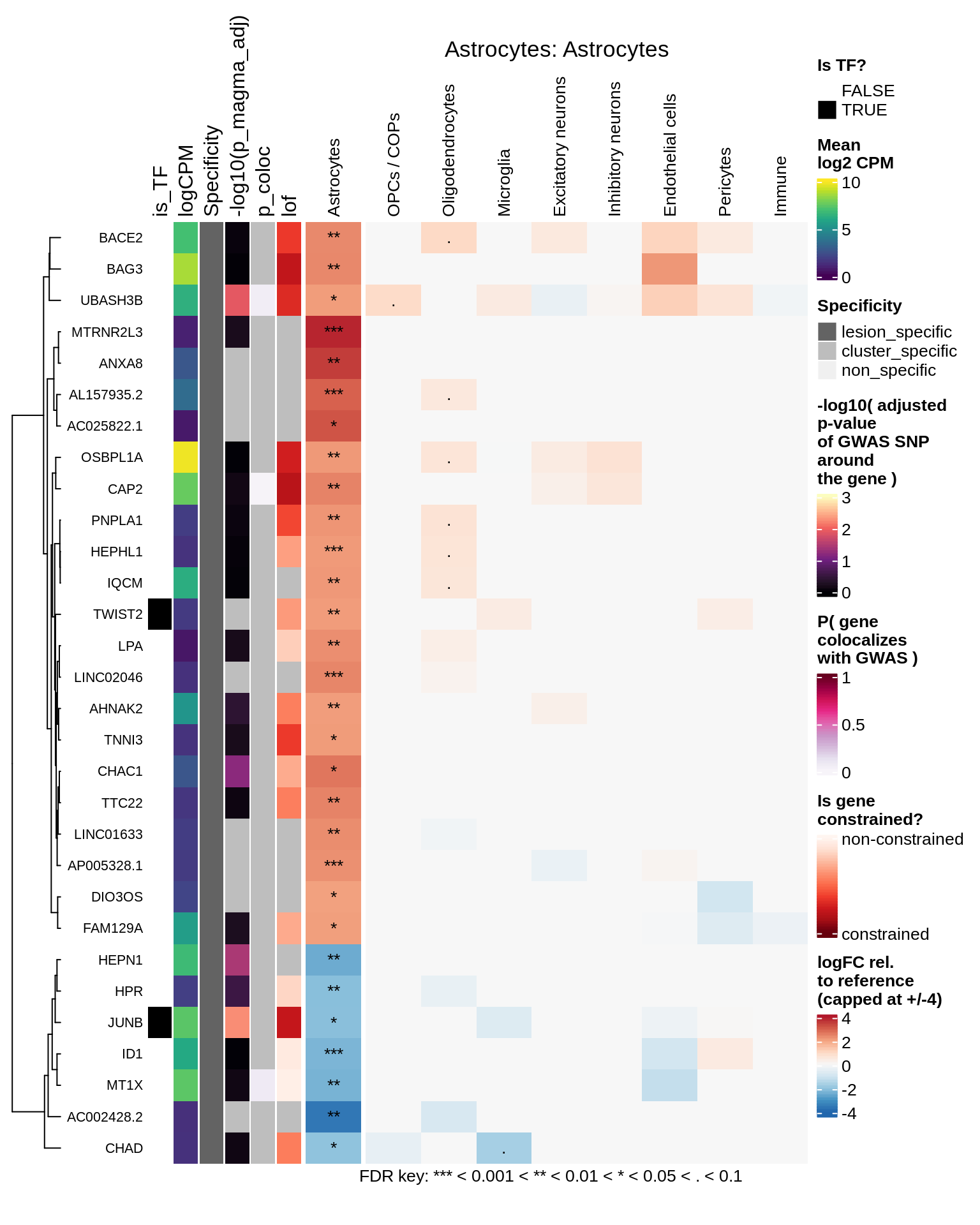
Microglia
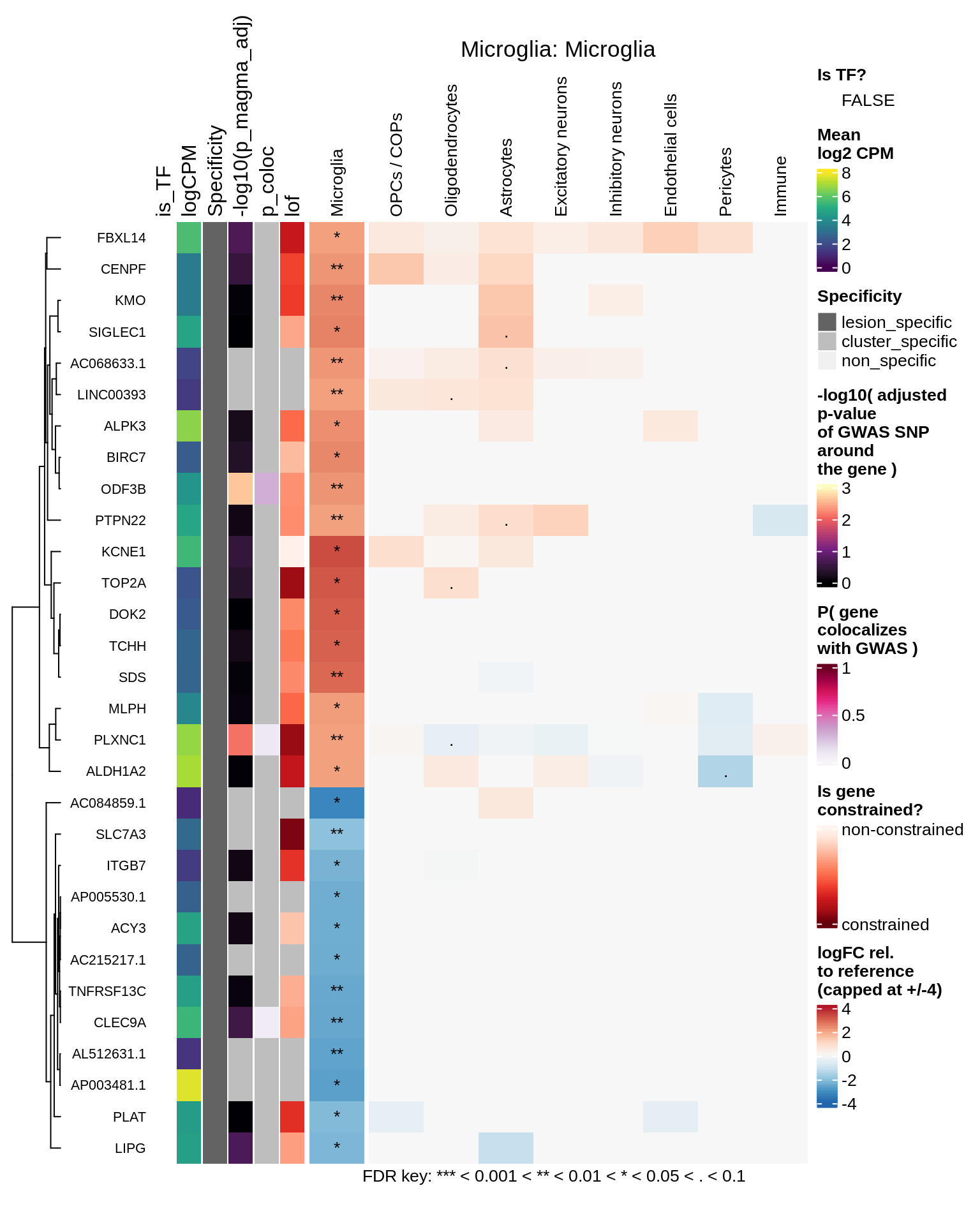
Excitatory neurons
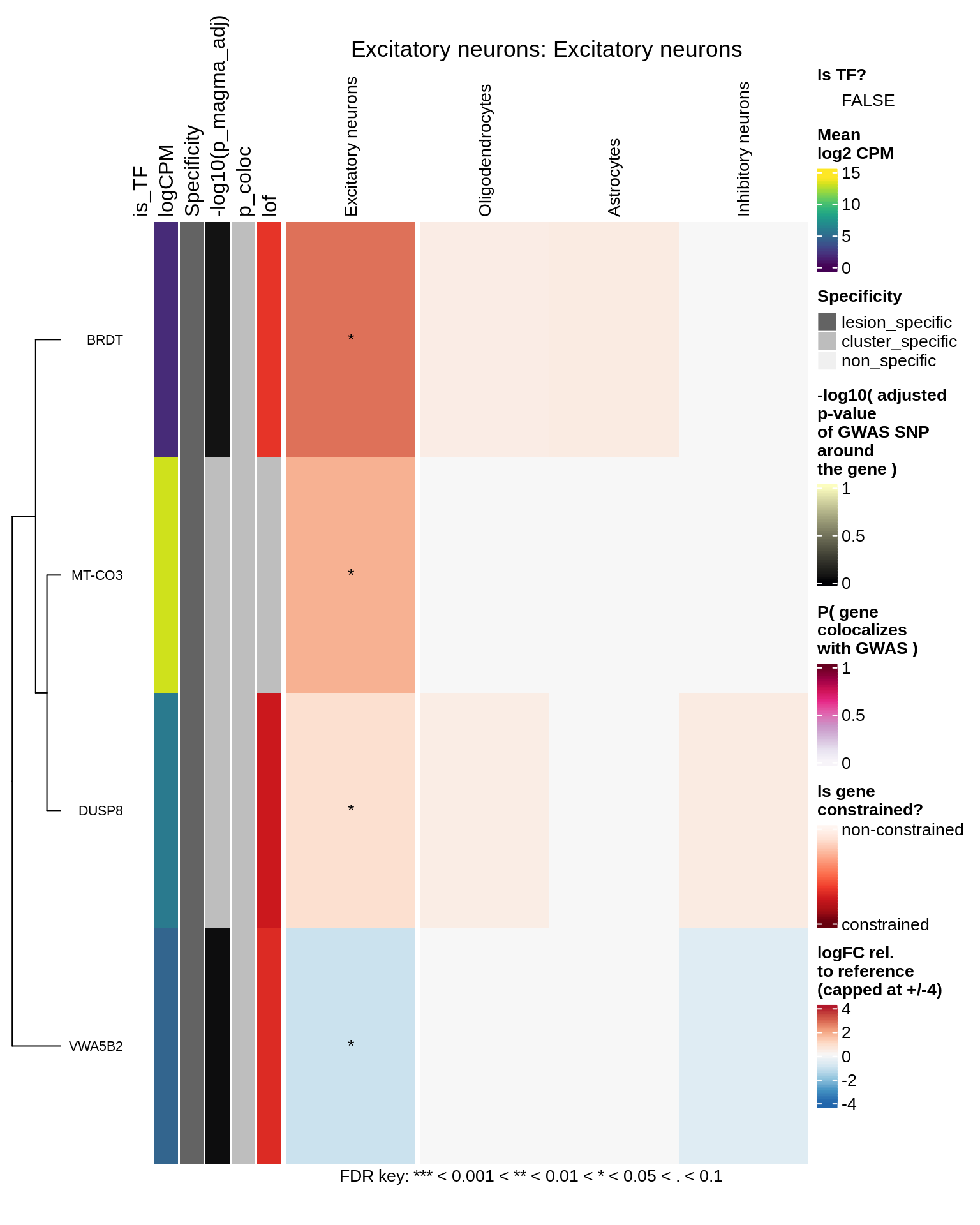
Inhibitory neurons
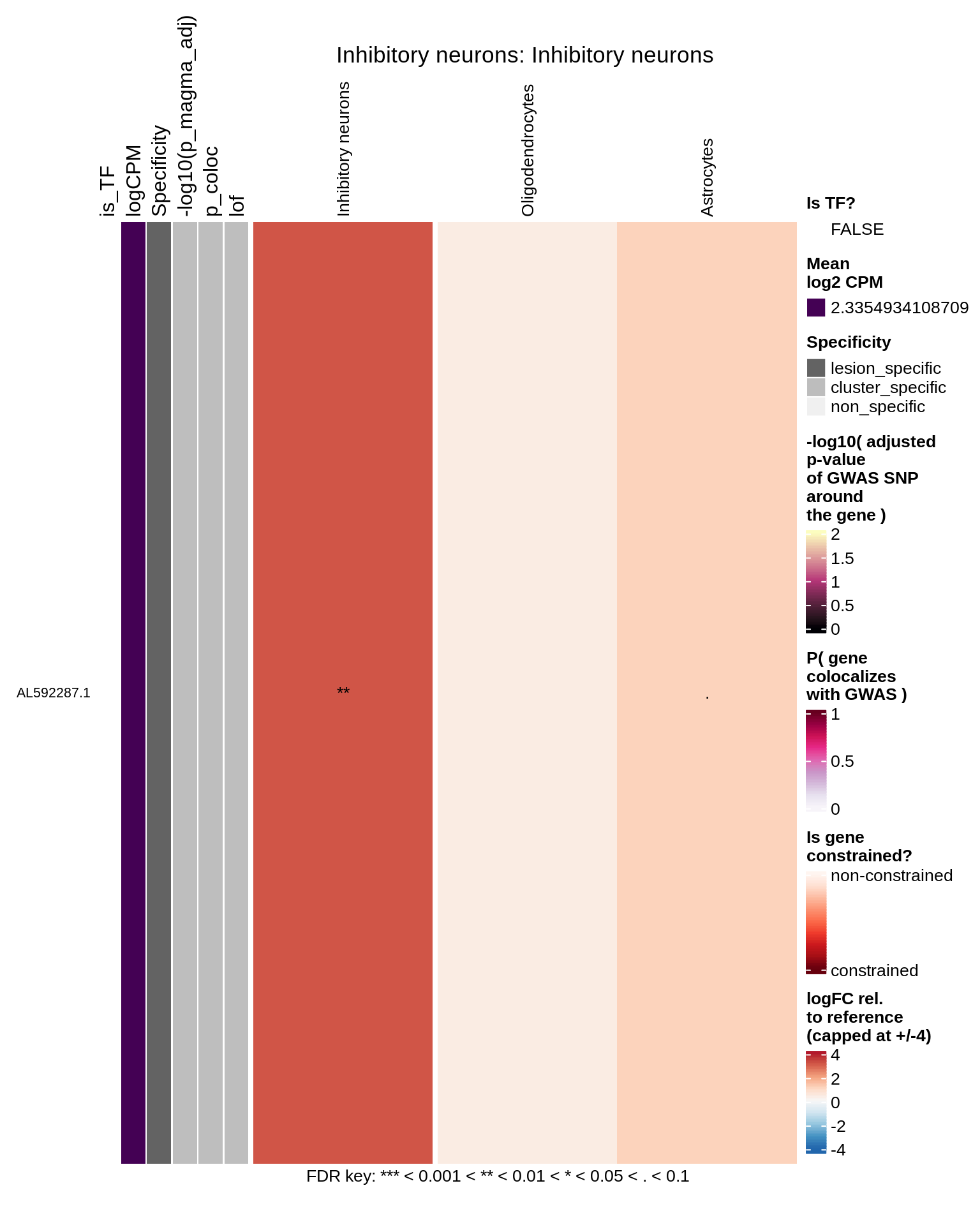
Endothelial cells
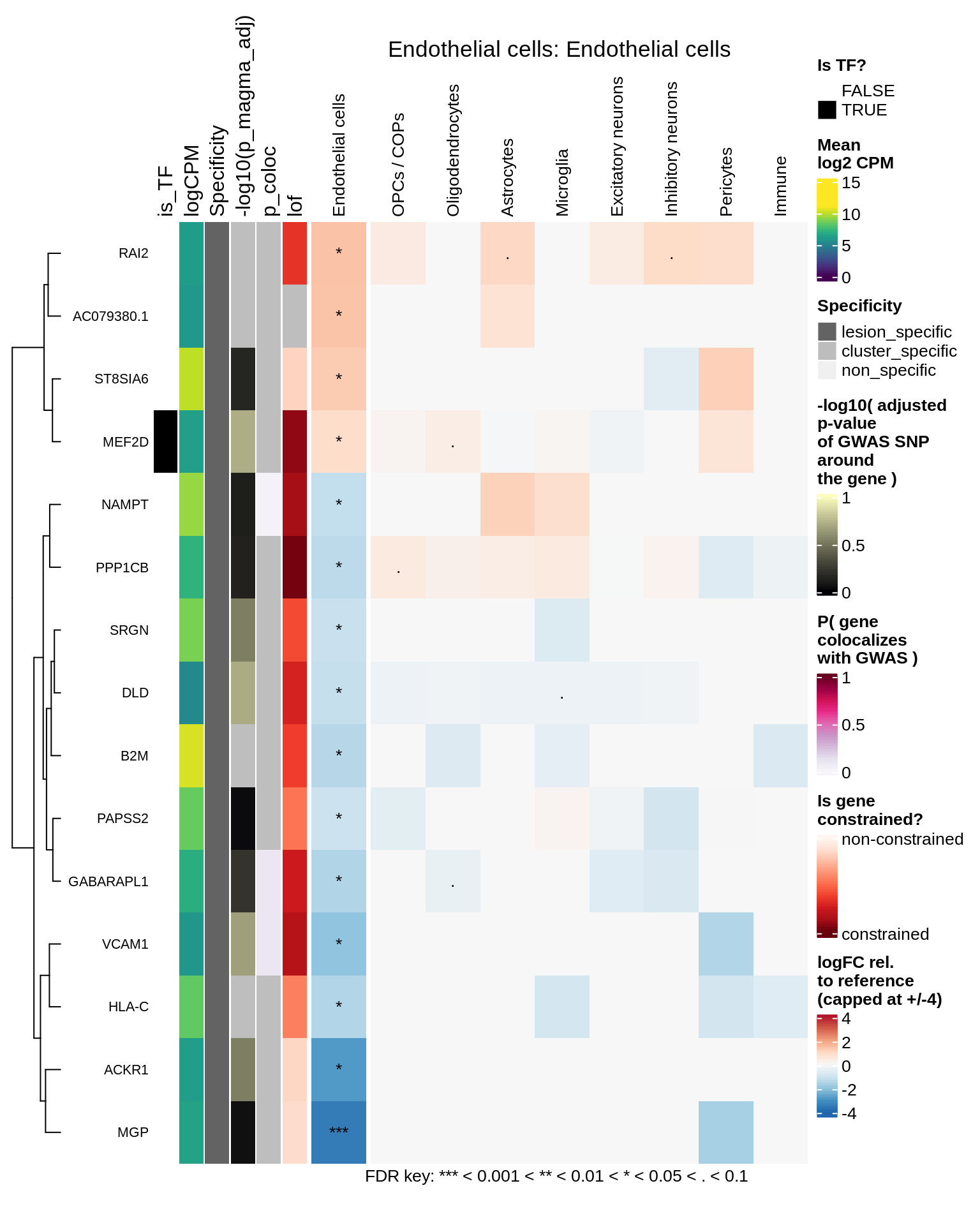
Pericytes
how_unique = 'non_specific'
cat('# Top ', how_unique, ' genes, ', this_type,
' variables{.tabset}\n', sep='')Top non_specific genes, test variables
for (cl in levels(top_dt$cluster_id)) {
tmp_dt = top_dt[ unique_var %in% unique_ls[[how_unique]] &
cluster_id == cl & test_var == this_var ]
tb = unique(tmp_dt$type_broad)
if ( nrow(tmp_dt) == 0 )
next
cat('## ', cl, '\n')
hm_obj = plot_top_genes_heatmap_fn_across_celltypes(tmp_dt, res_dt, labels_dt,
max_fc = 4, title = paste(tb, cl, sep=': '))
draw(hm_obj, padding = unit(c(0.5, 0.1, 0.1, 0.1), "in"))
.add_fdr_legend('logfc')
cat('\n\n')
}OPCs / COPs
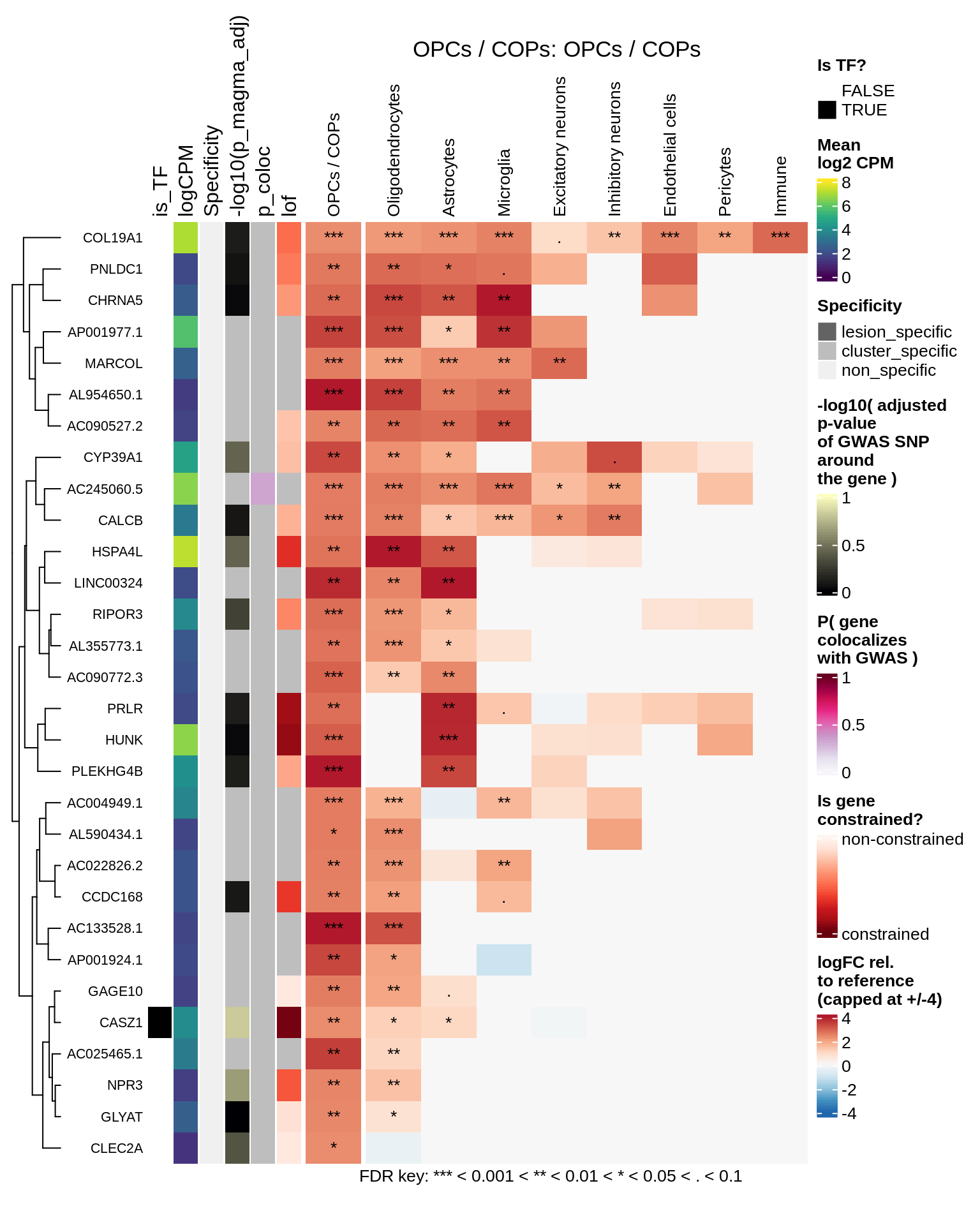
Oligodendrocytes
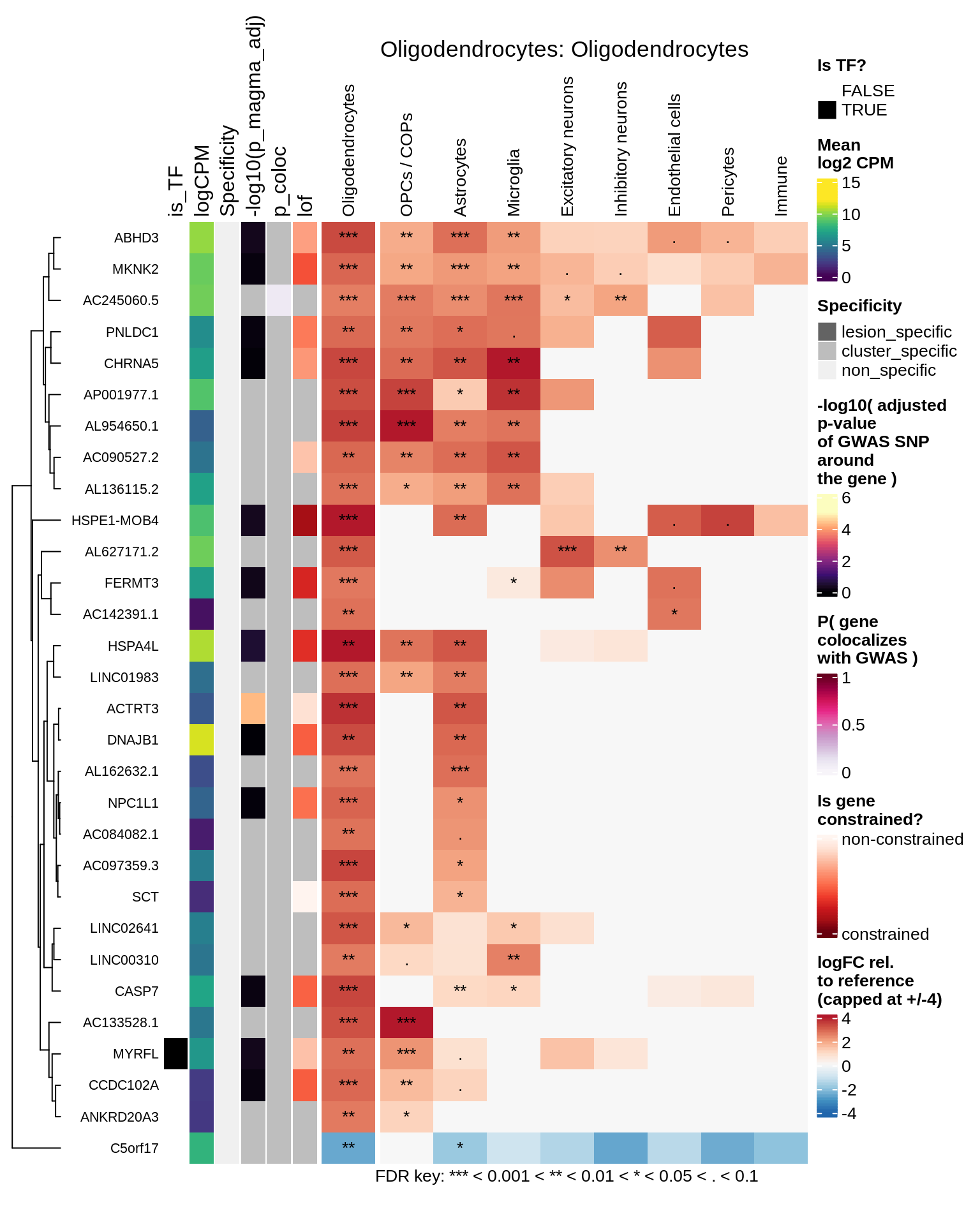
Astrocytes
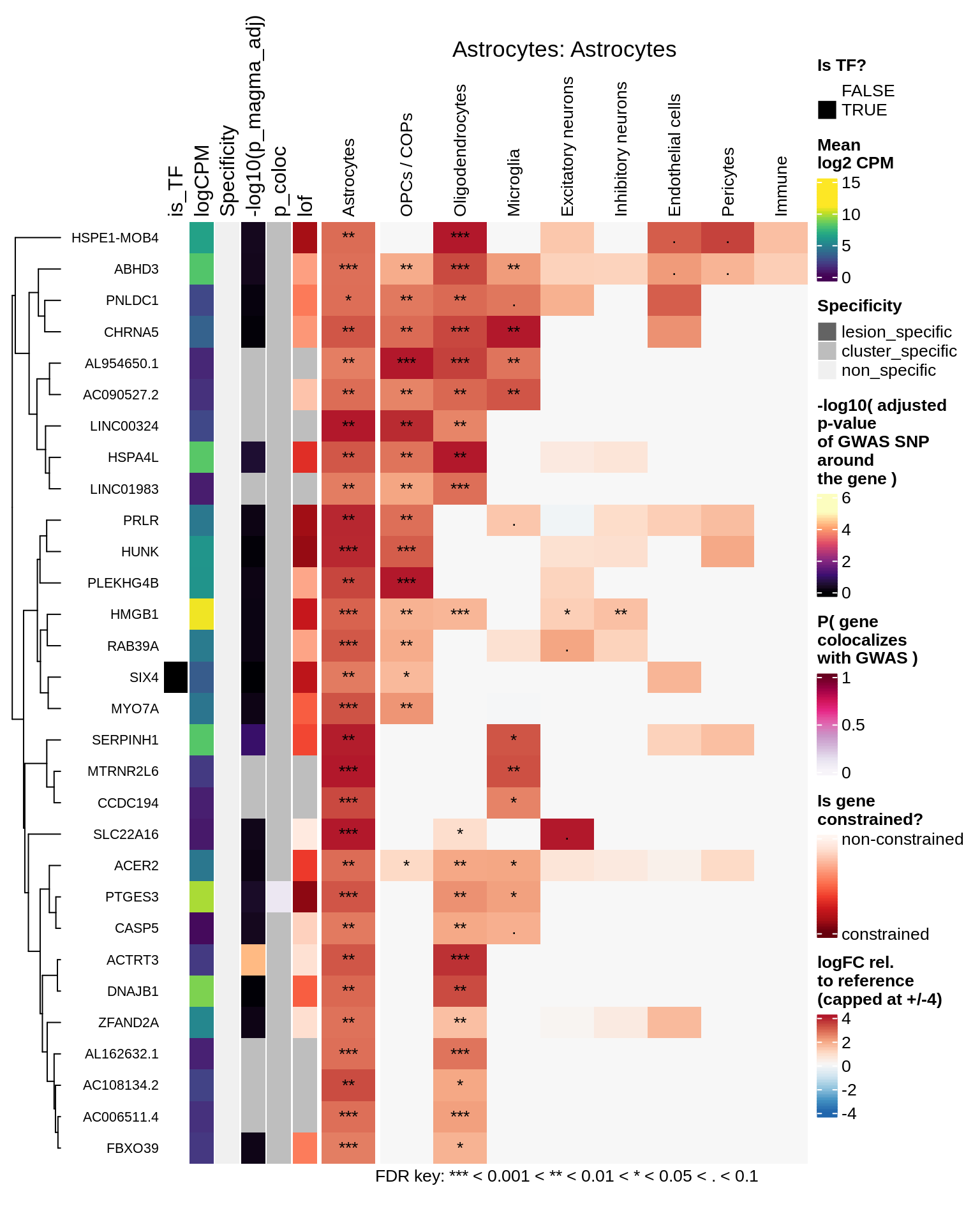
Microglia
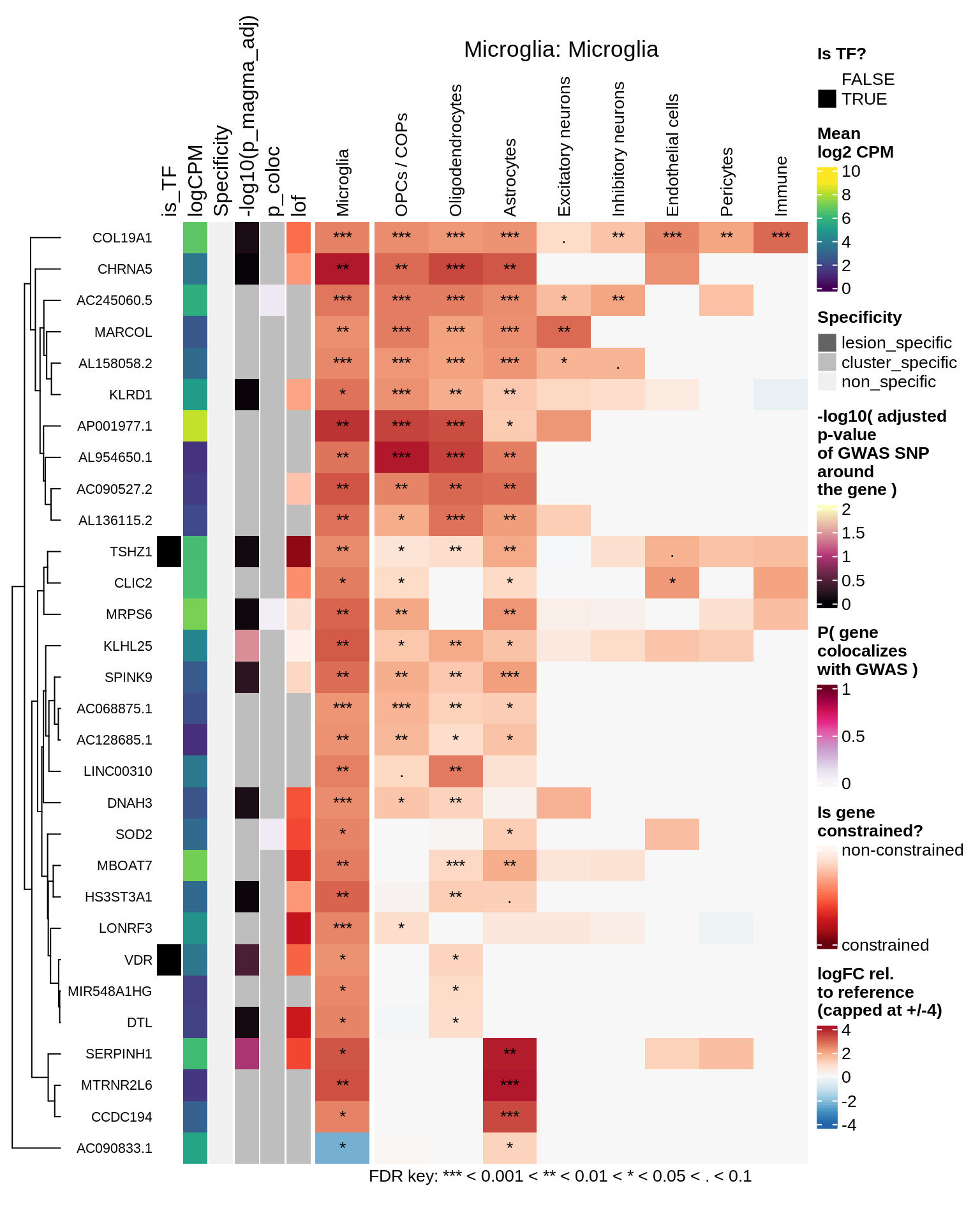
Excitatory neurons
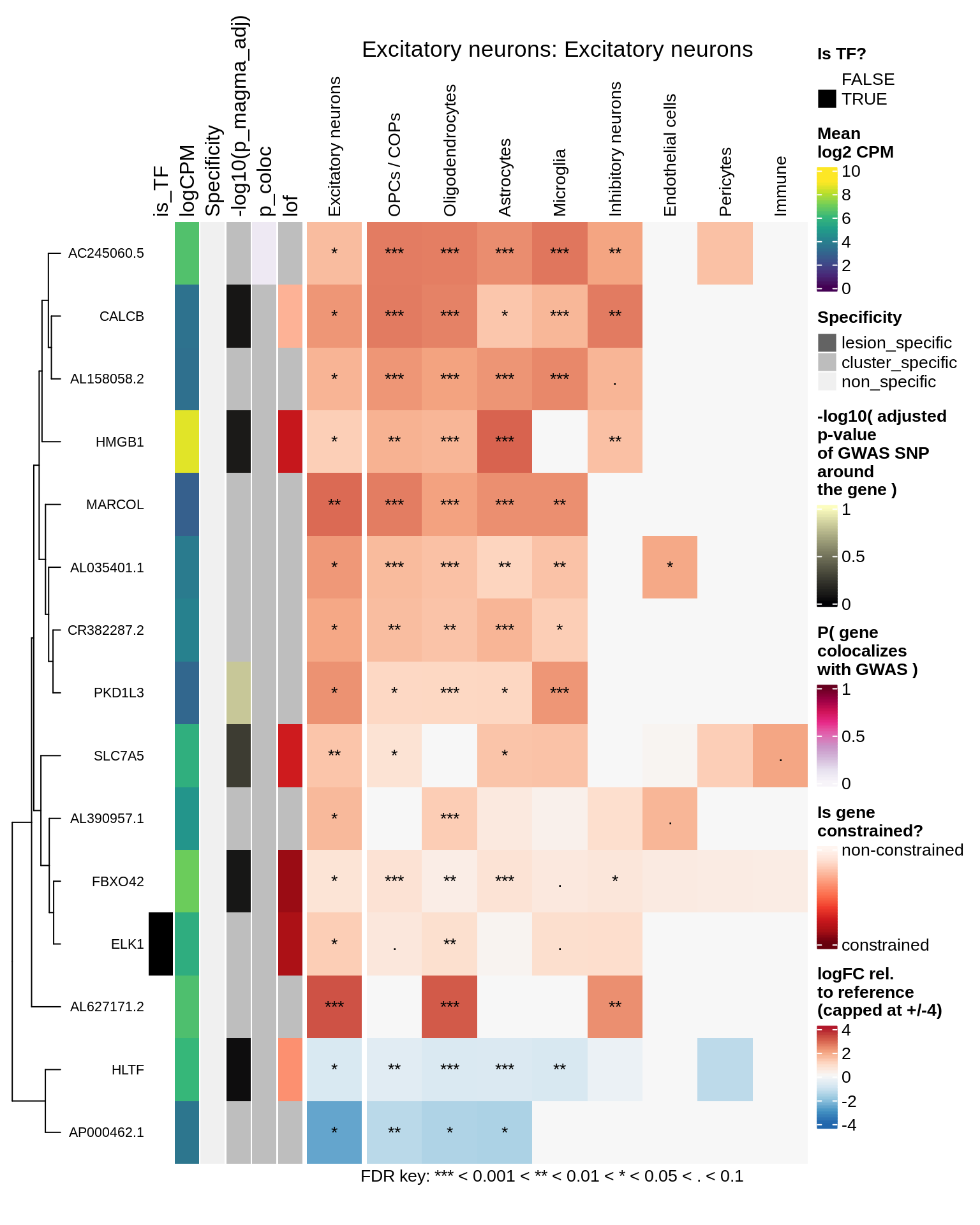
Inhibitory neurons
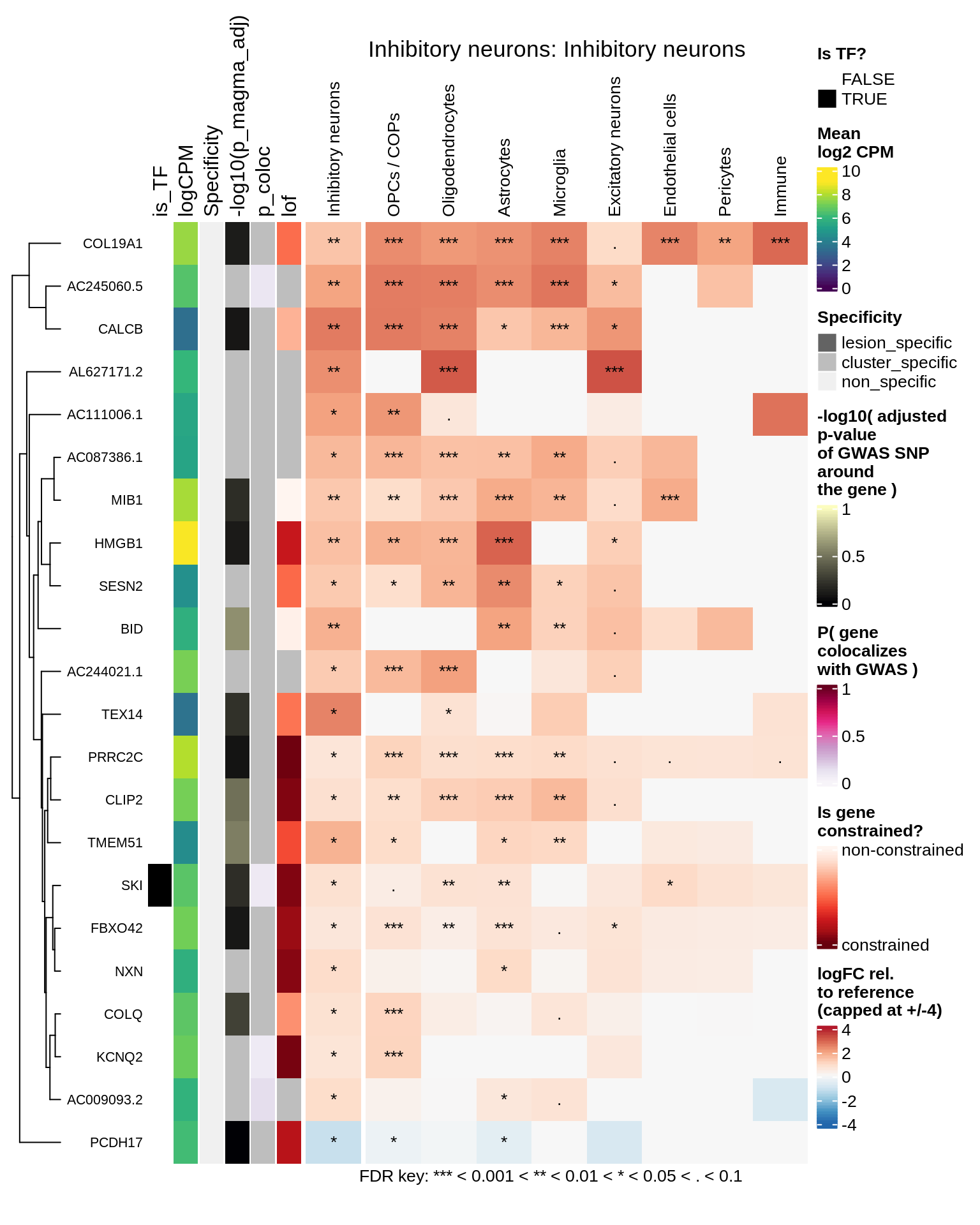
Endothelial cells
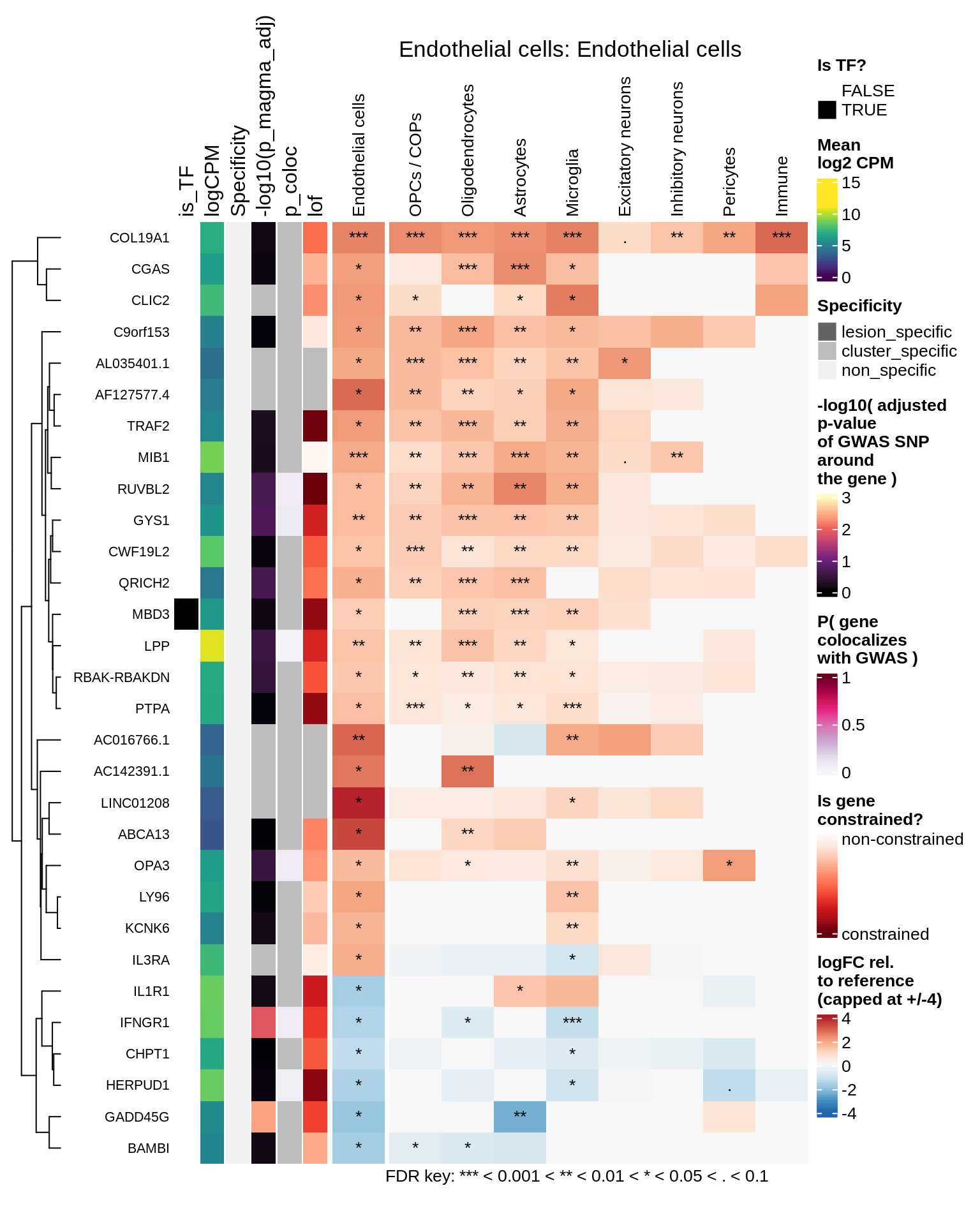
Pericytes
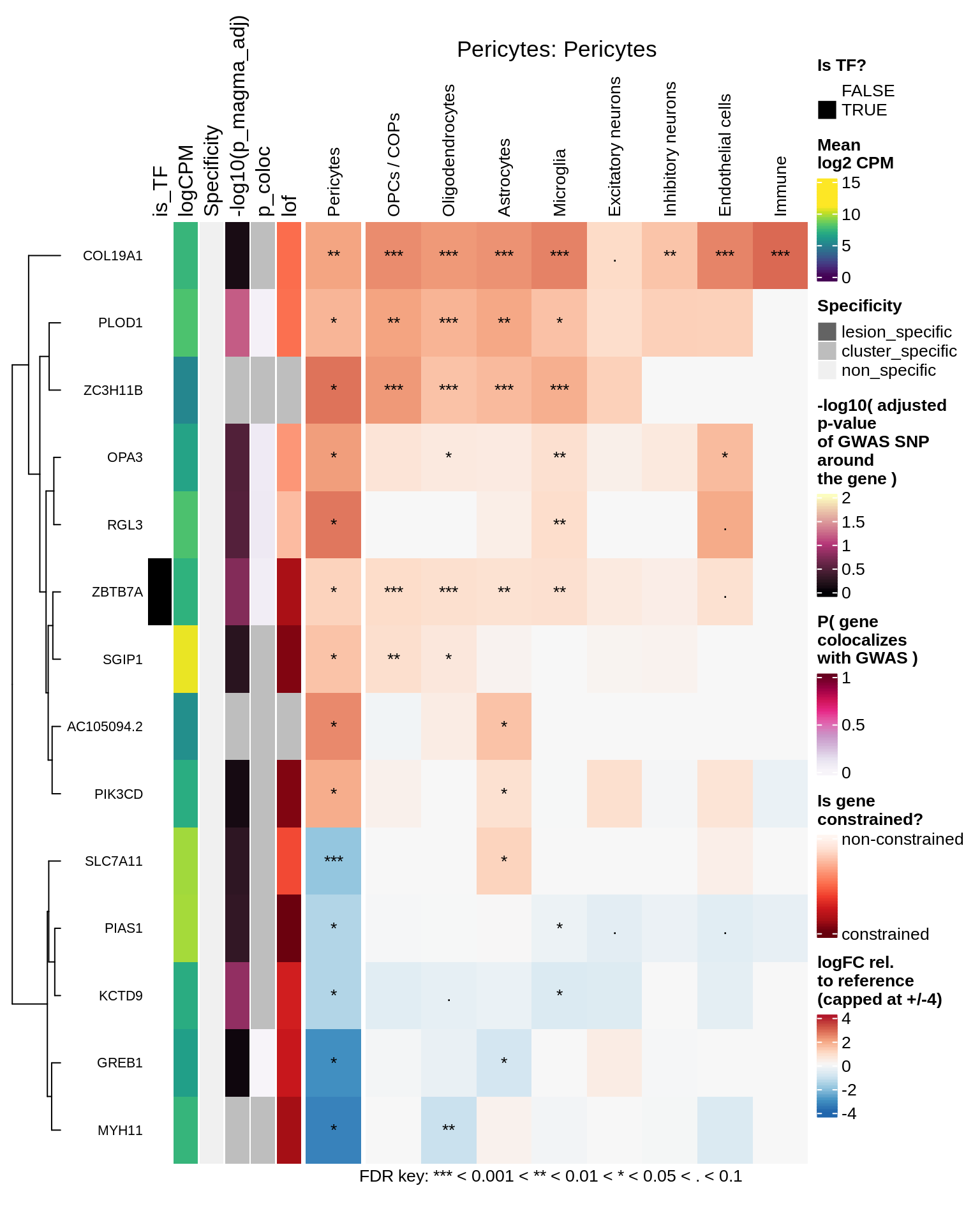
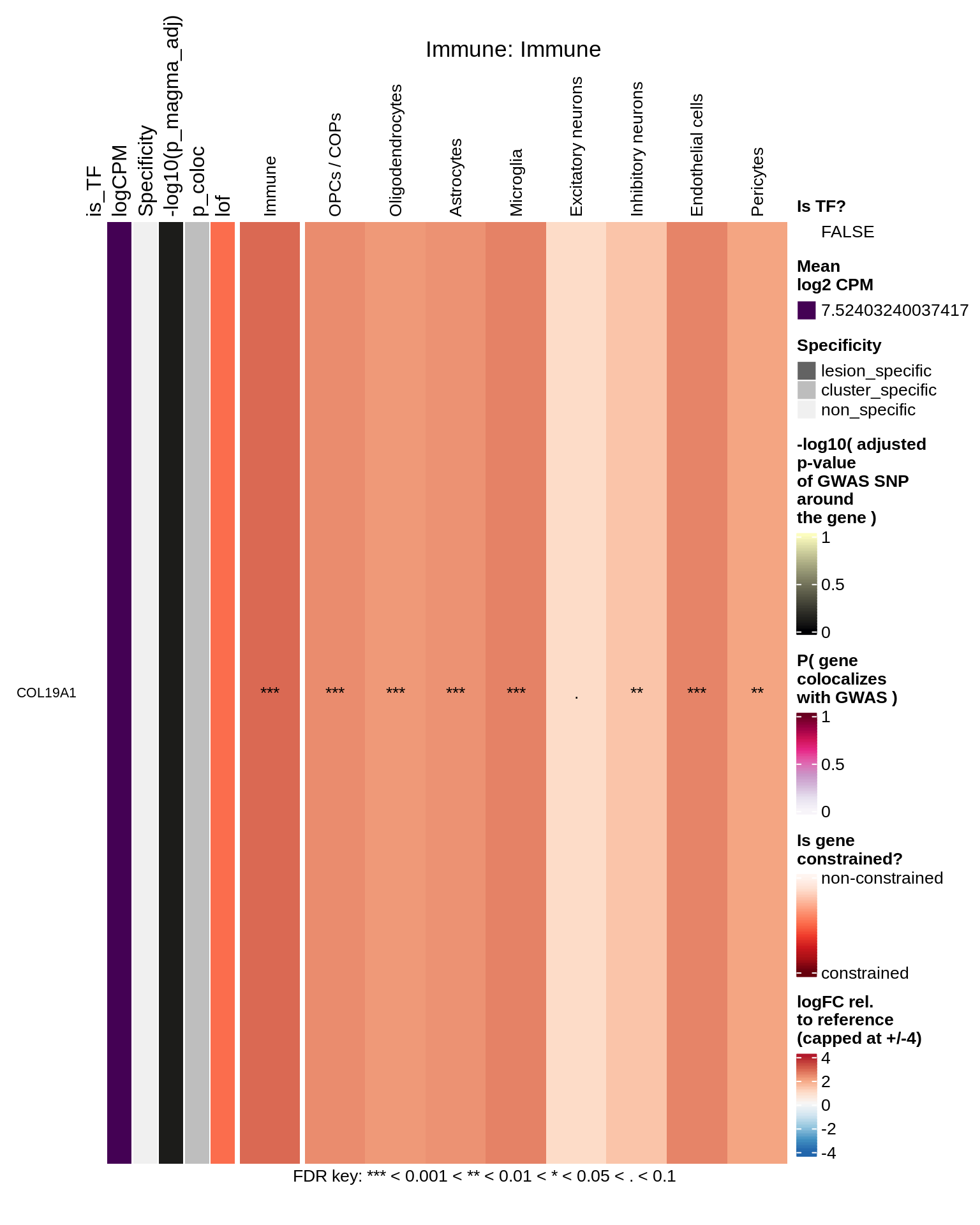
Immune
this_type = 'test'
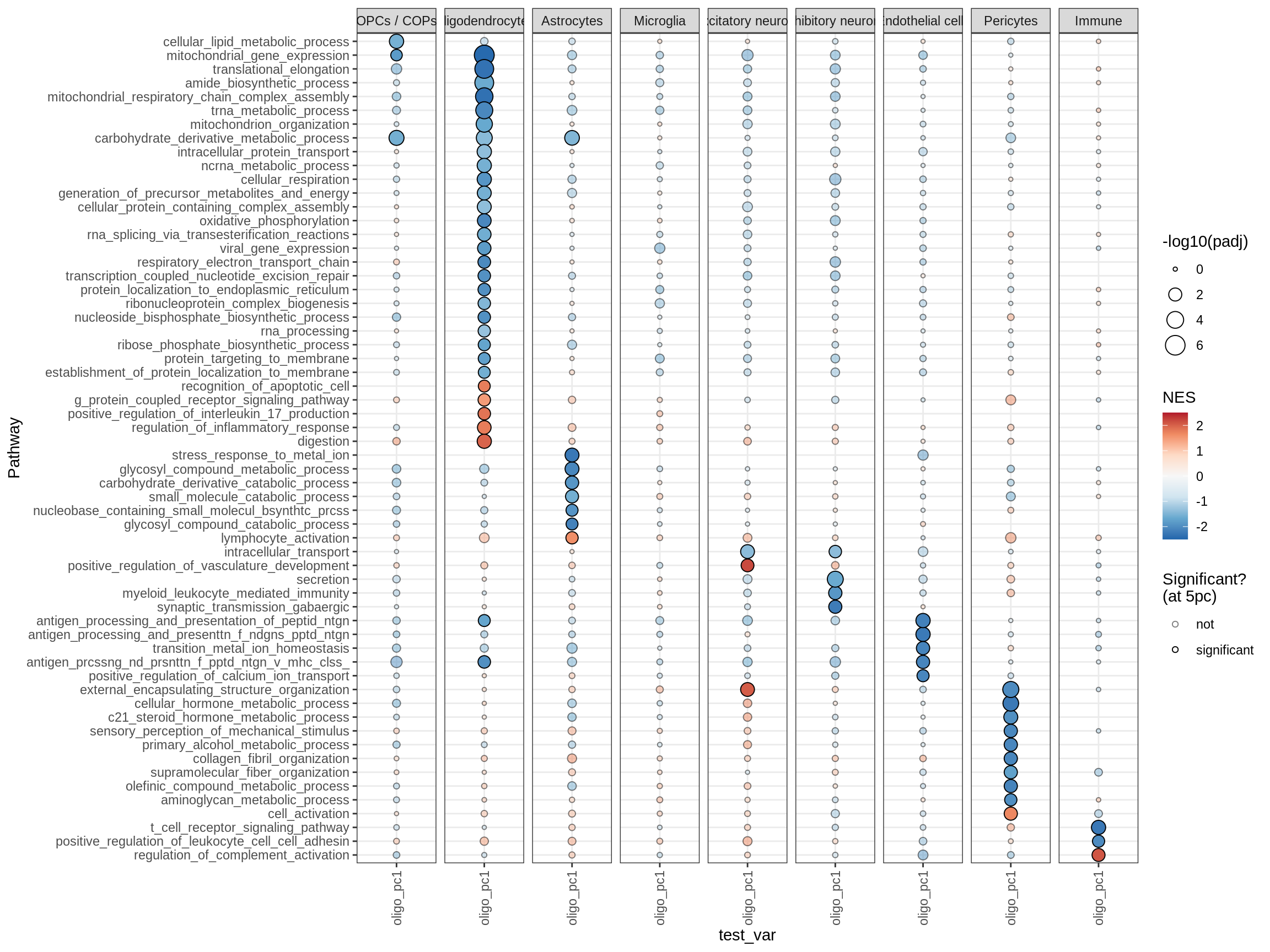
cat('# GSEA results for ', this_type, ' variables{.tabset}\n', sep = '')GSEA results for test variables
for (p in names(paths_list)) {
# restrict to relevant GO terms
cat('## ', p, '\n', sep='')
dt = fgsea_list[[p]] %>% .[var_type == this_type]
if (nrow(dt[ padj < fgsea_cut ]) == 0)
next
# plot
print(plot_gsea_dotplot(dt, labels_dt, top_n_paths = 60))
cat('\n\n')
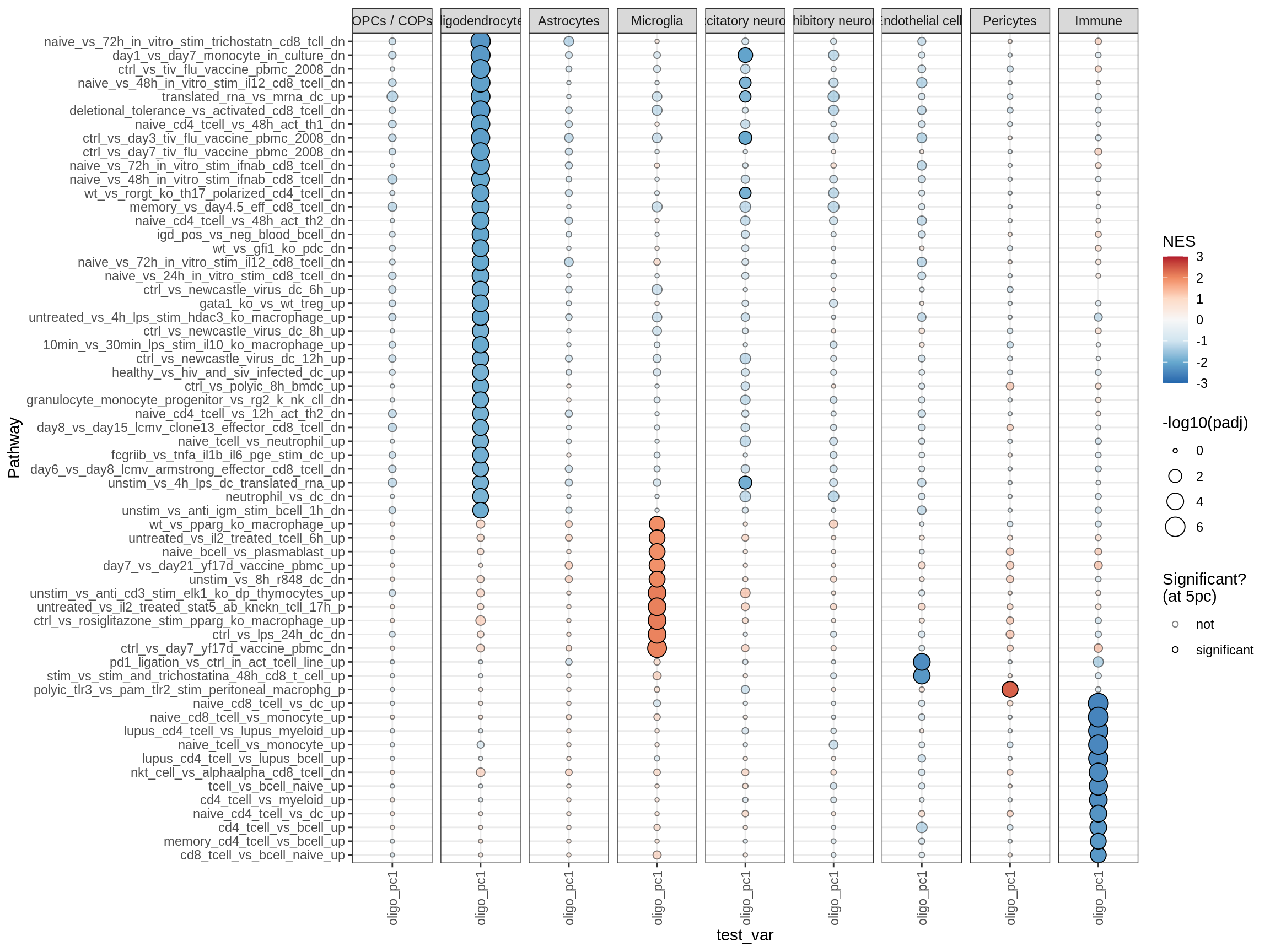
}go_bp
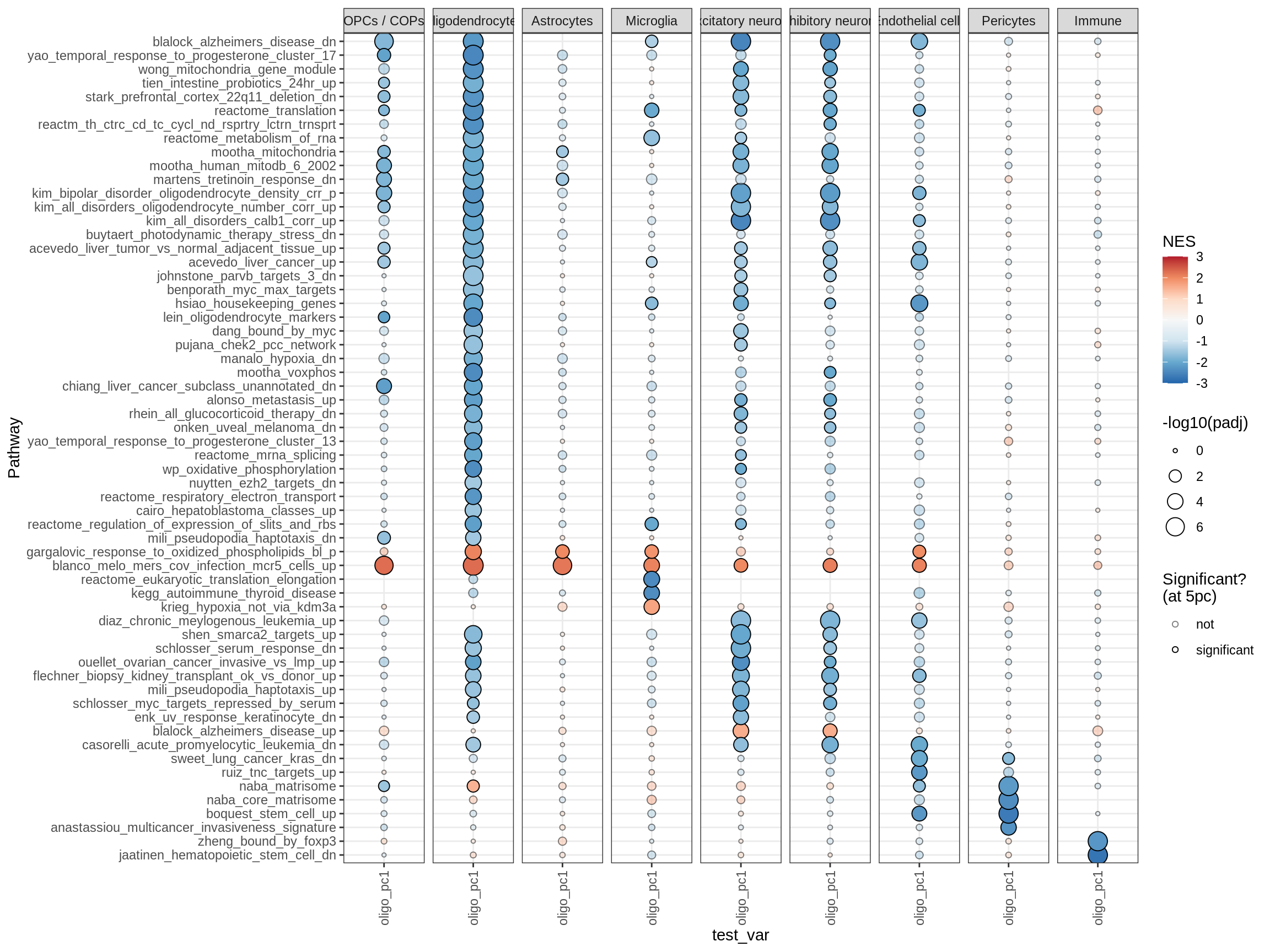
hallmark
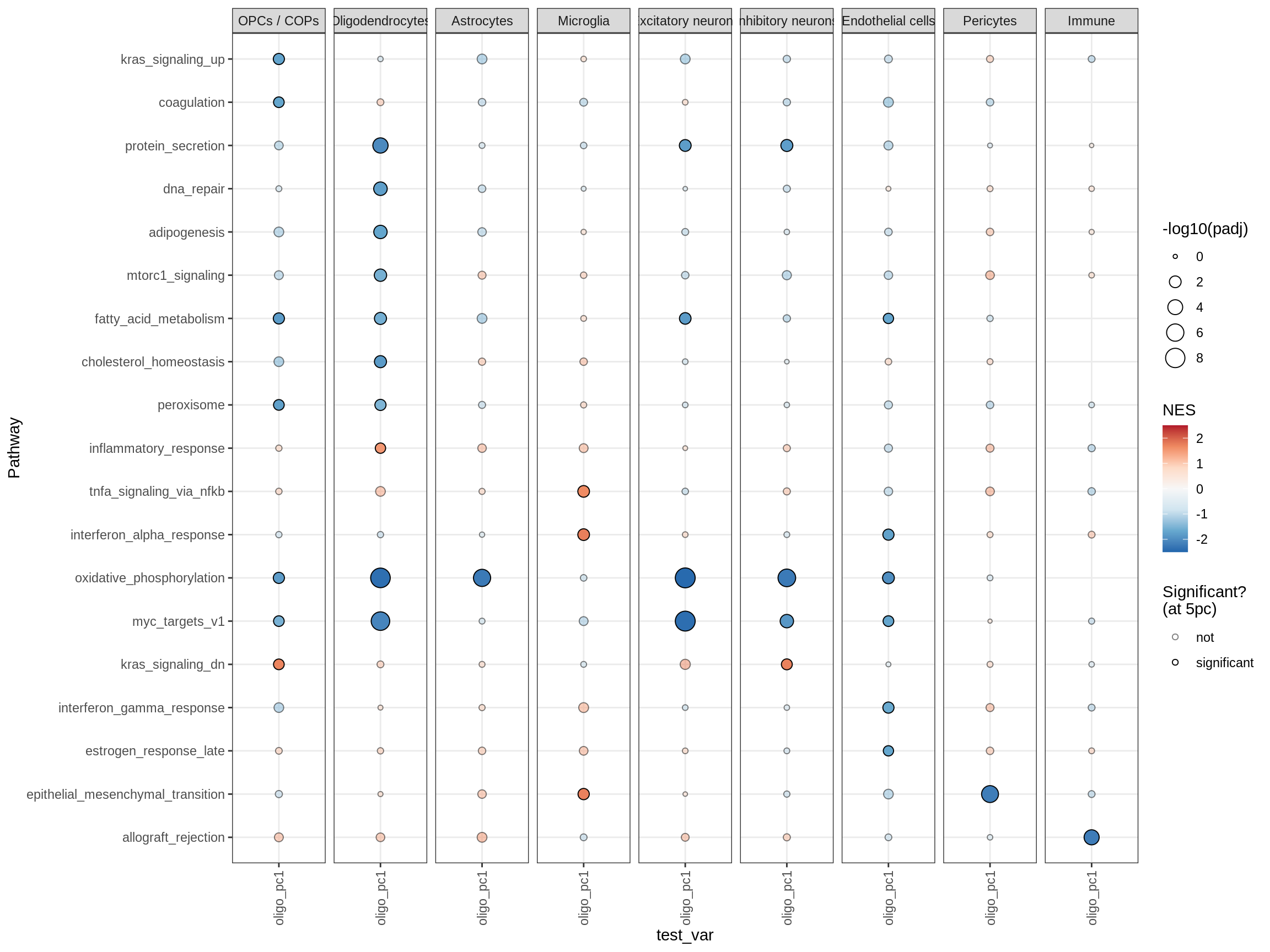
TFs
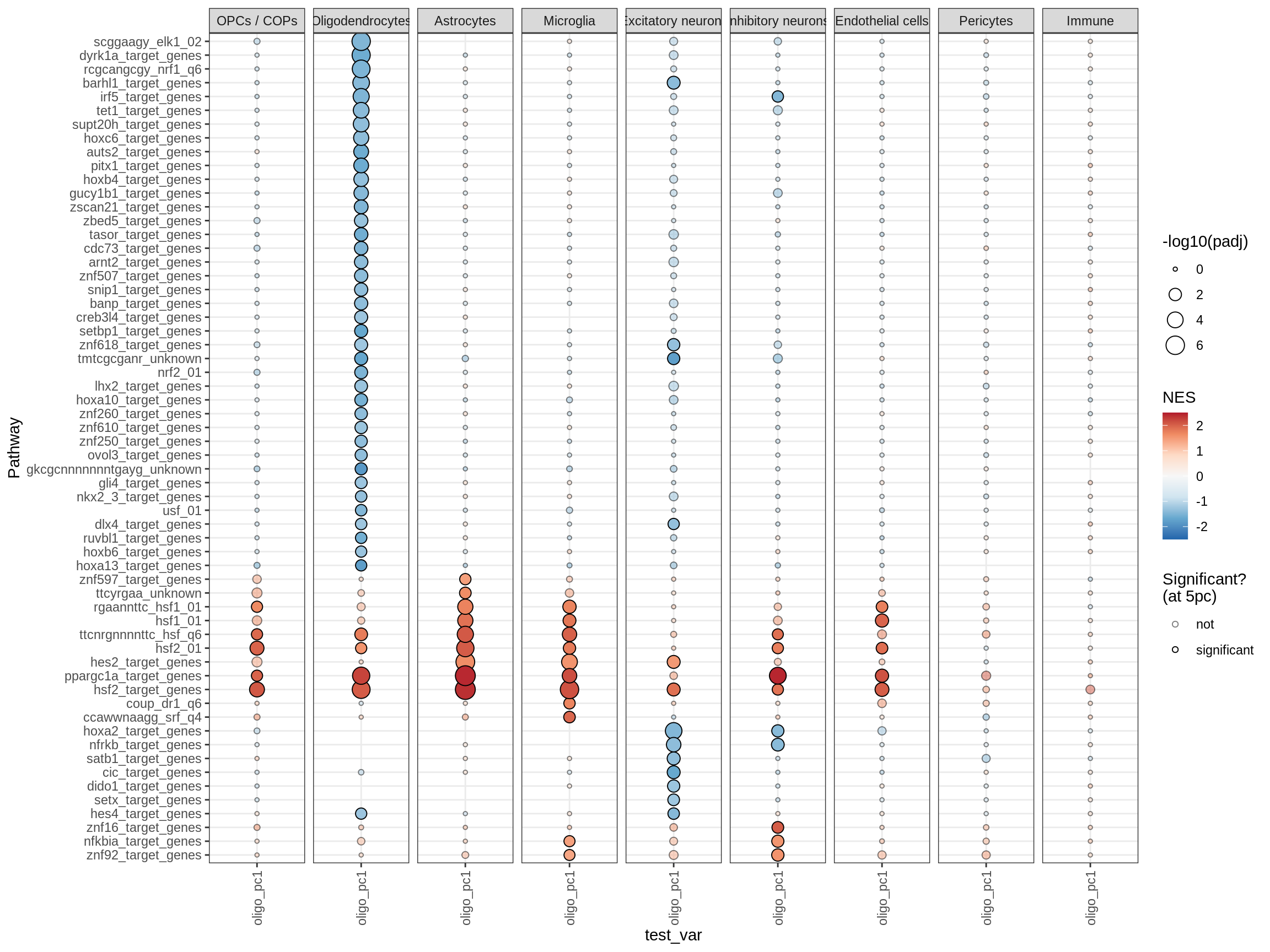
celltype
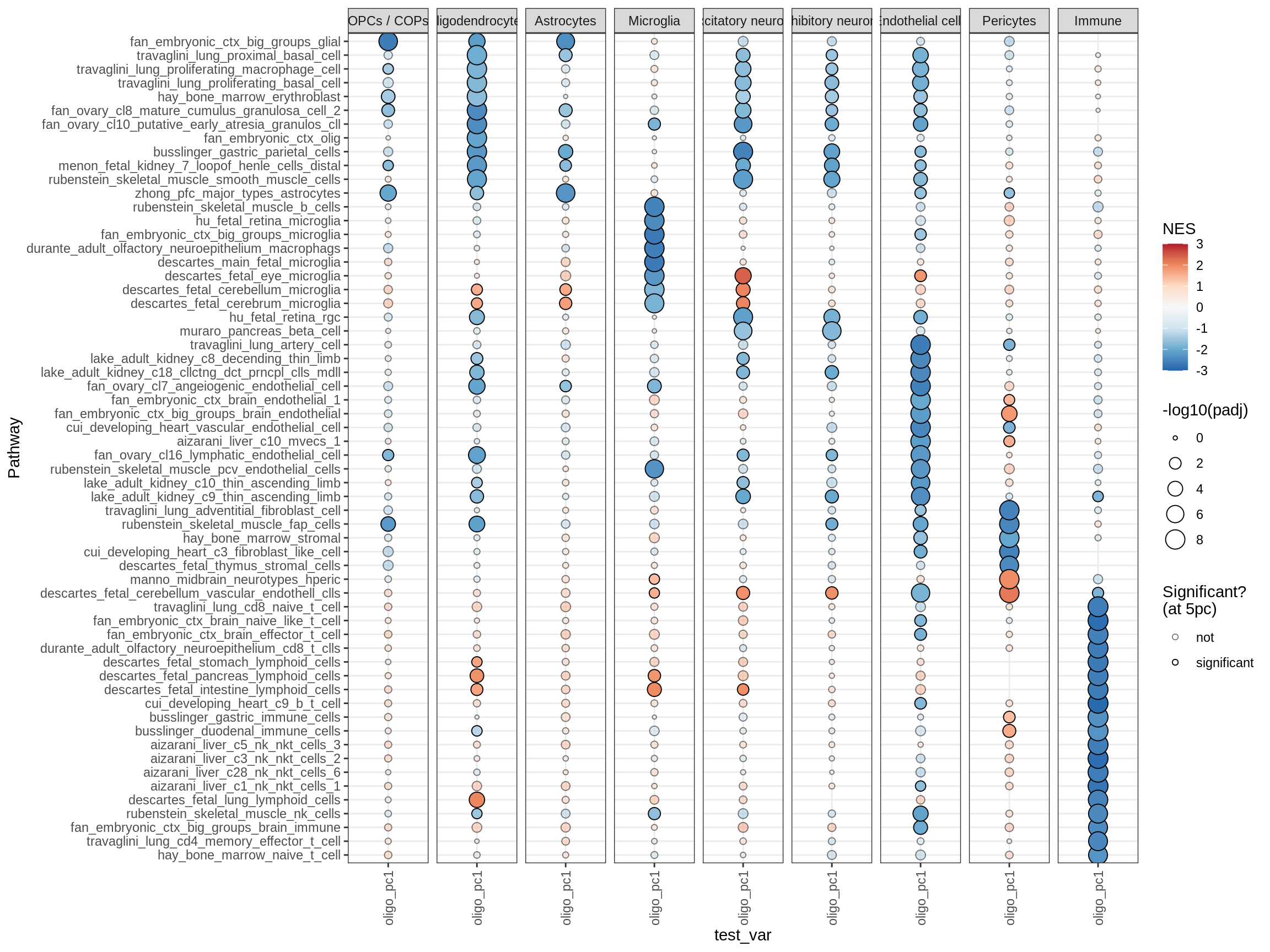
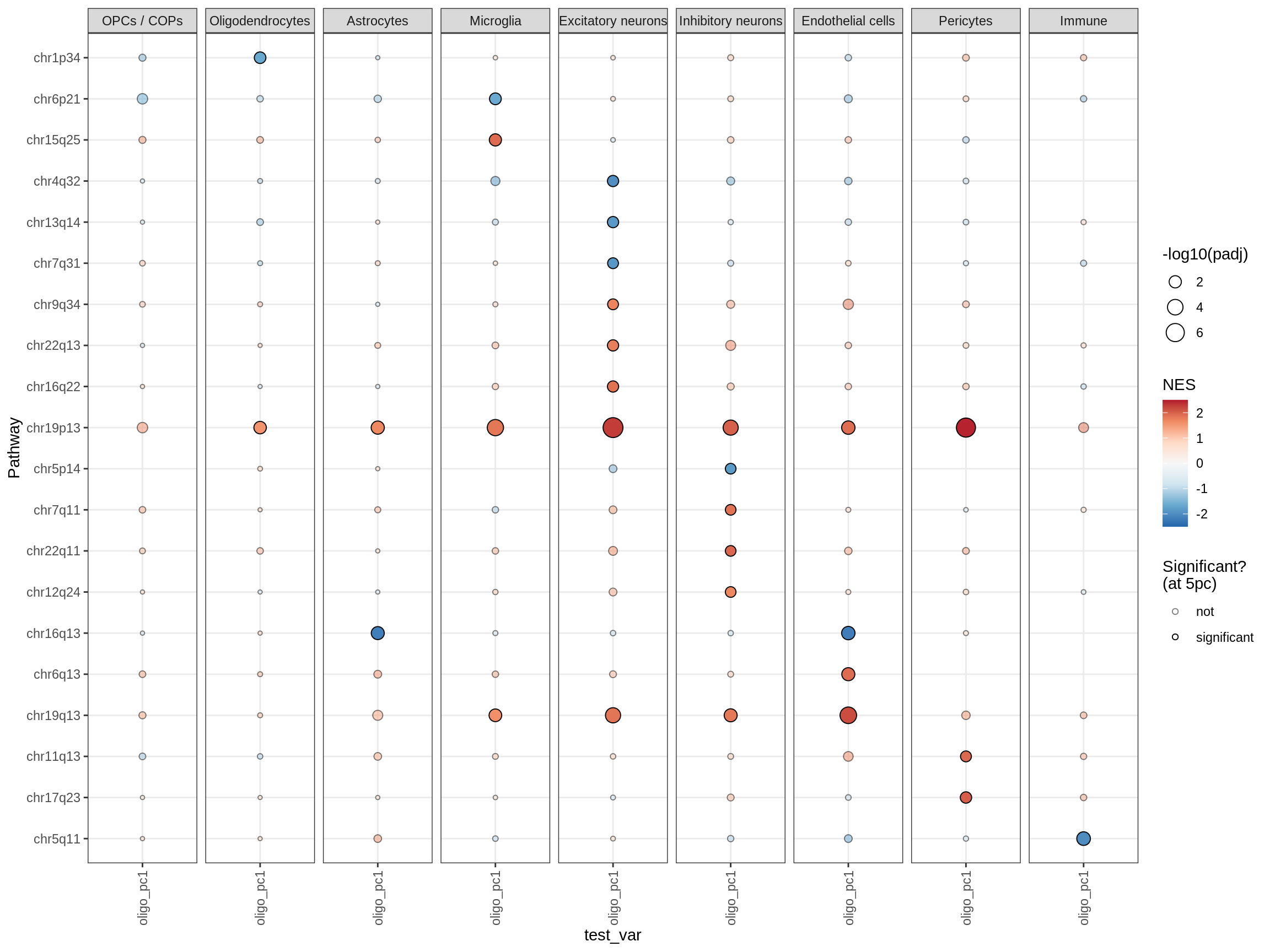
hpo
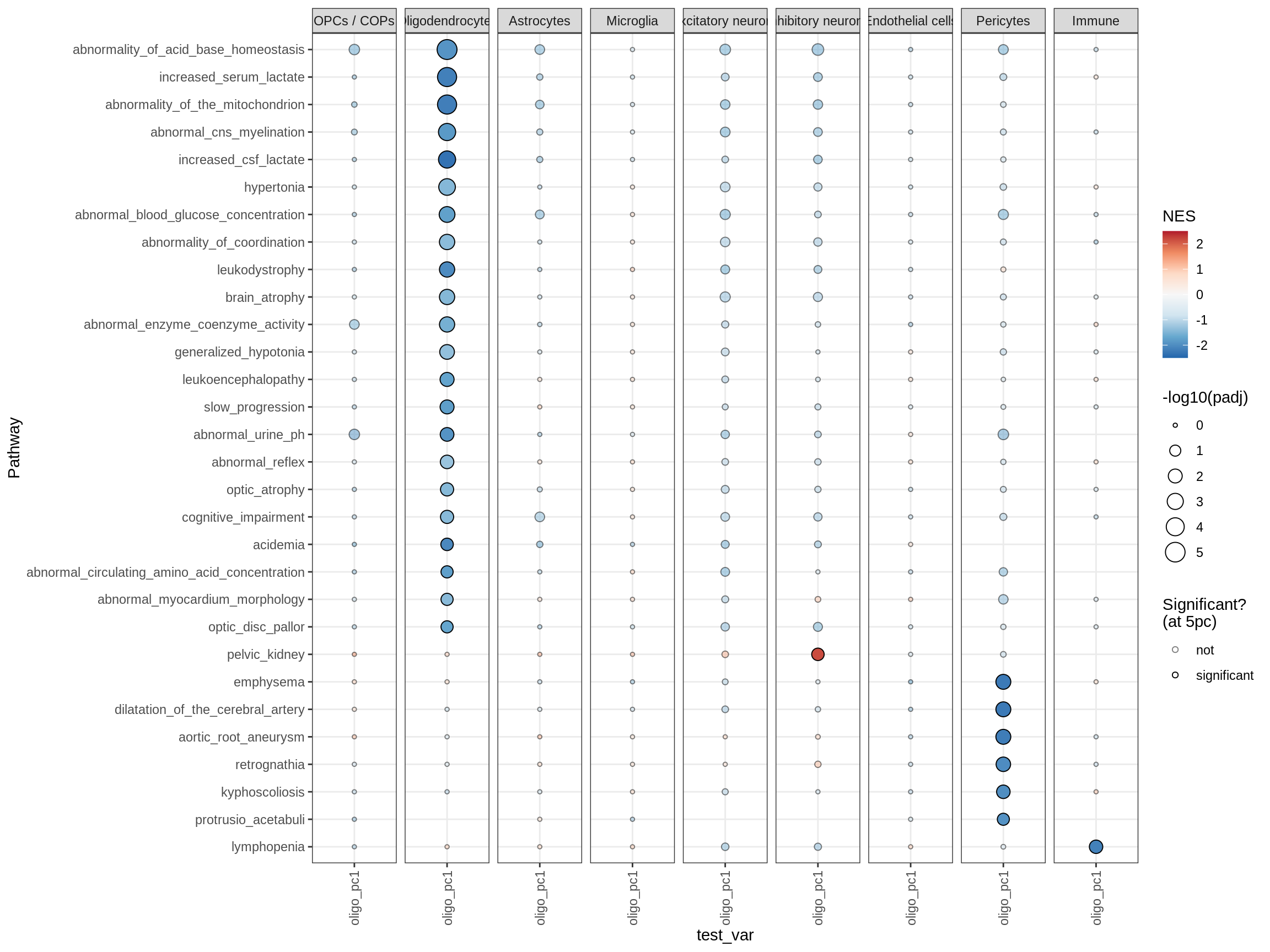
mirs
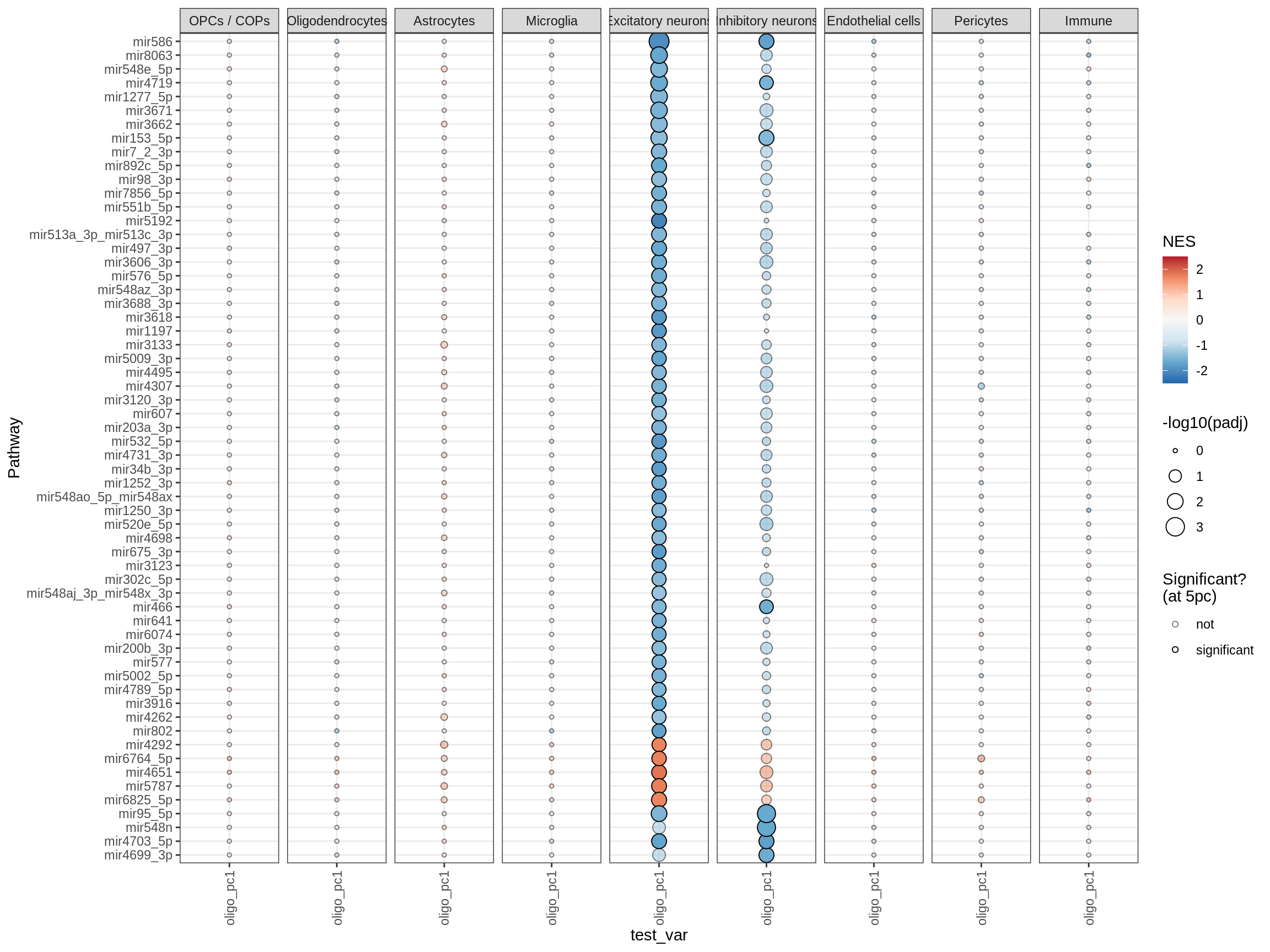
curated
immune
position
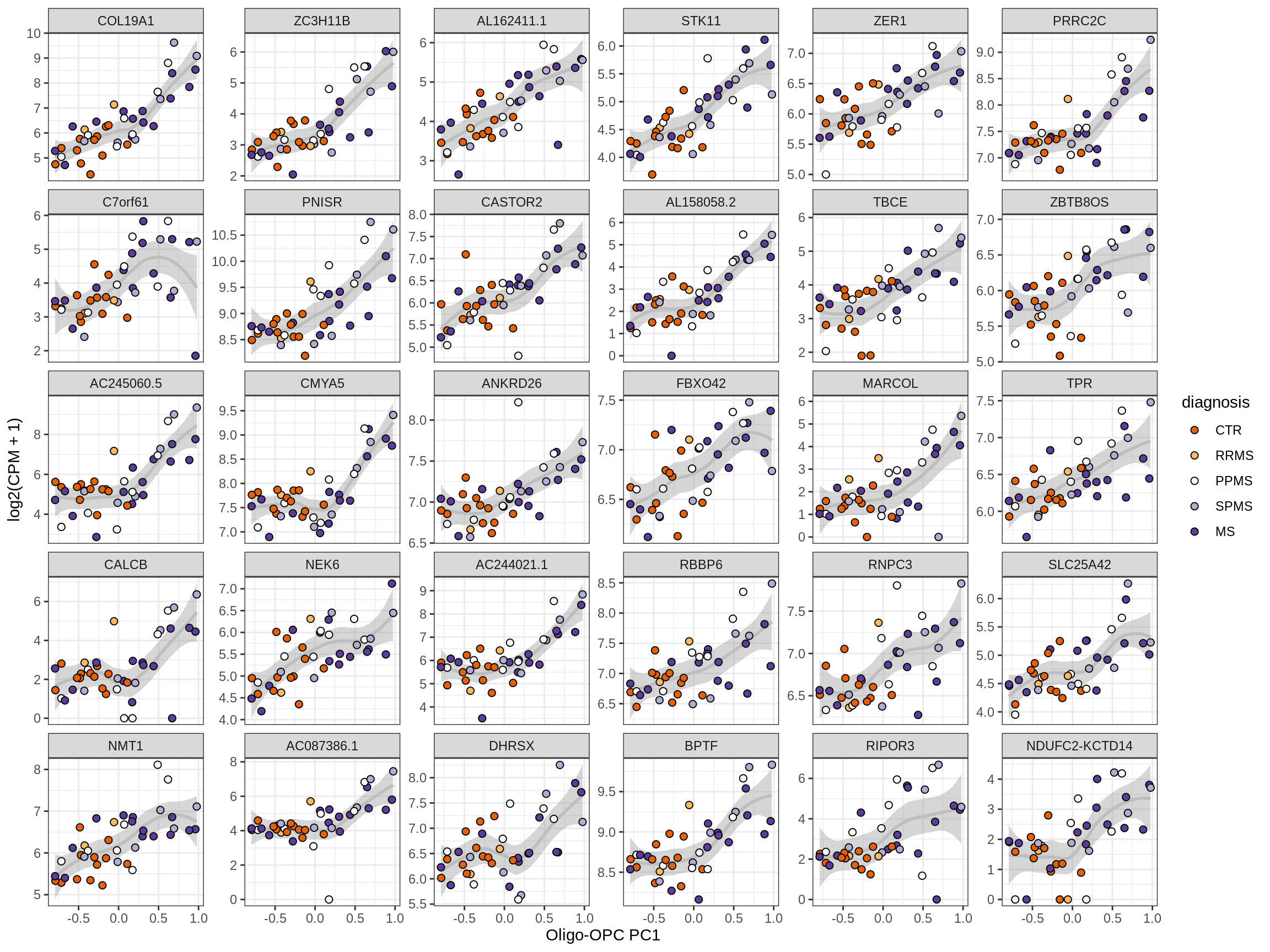
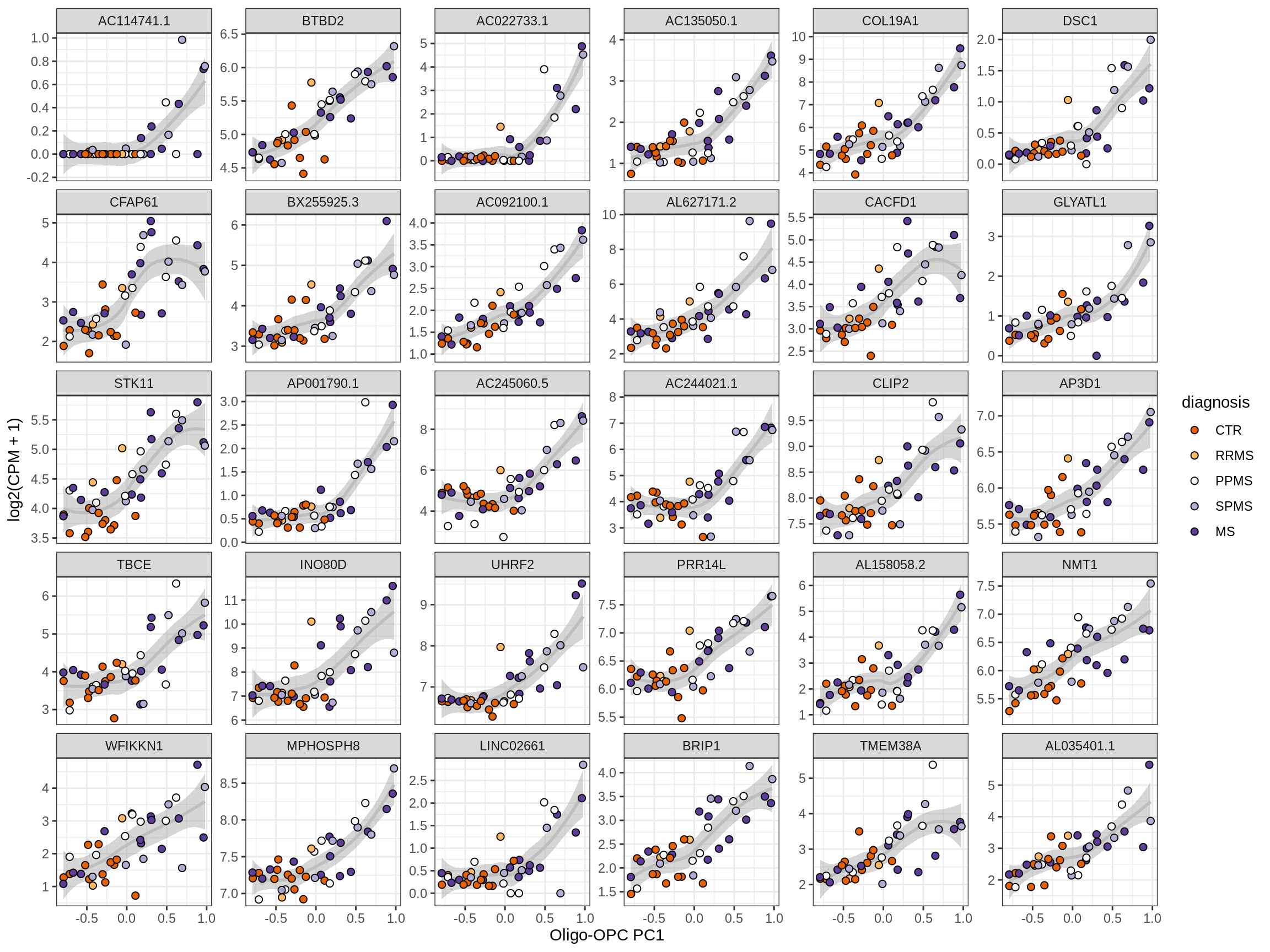
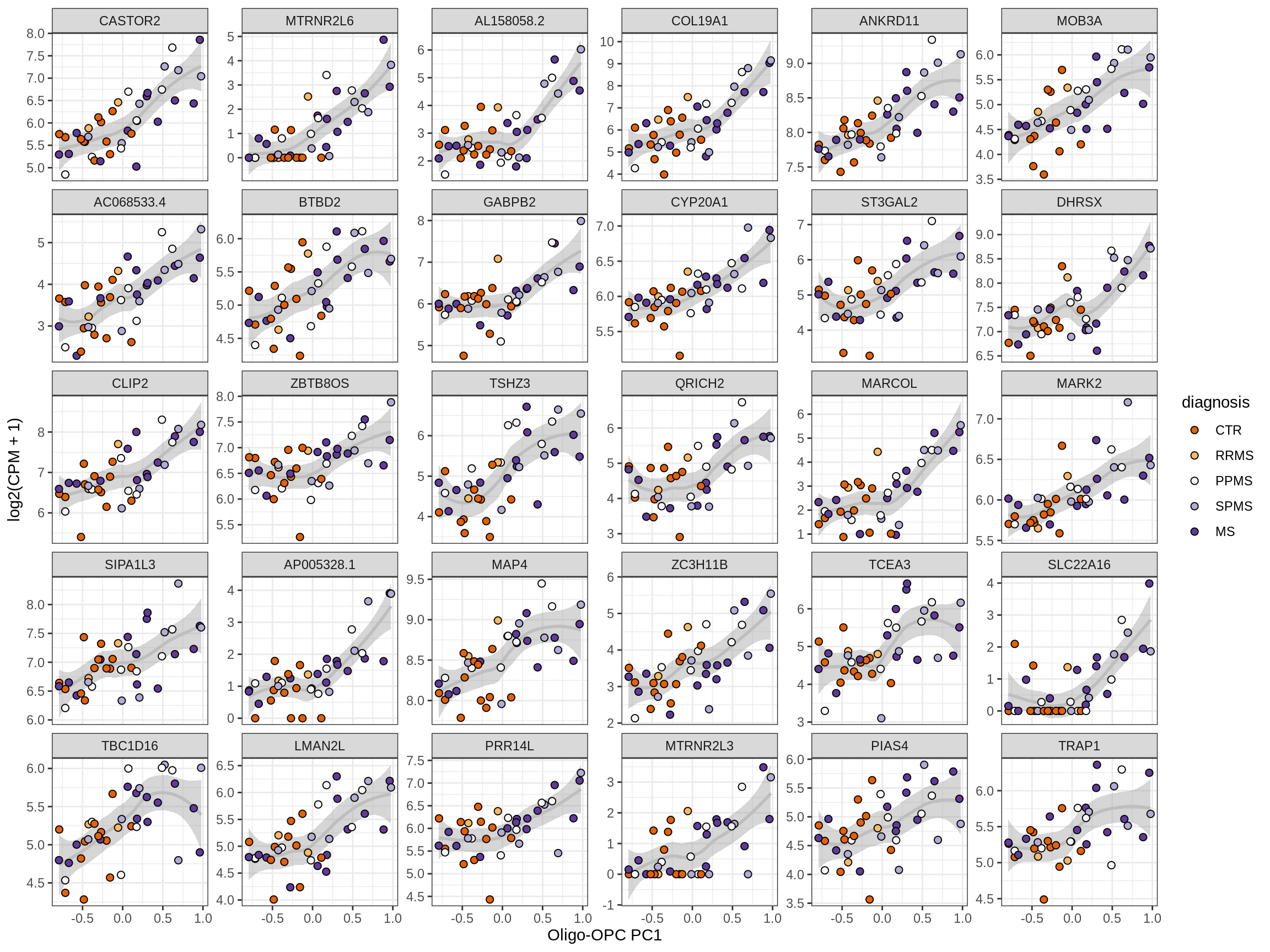
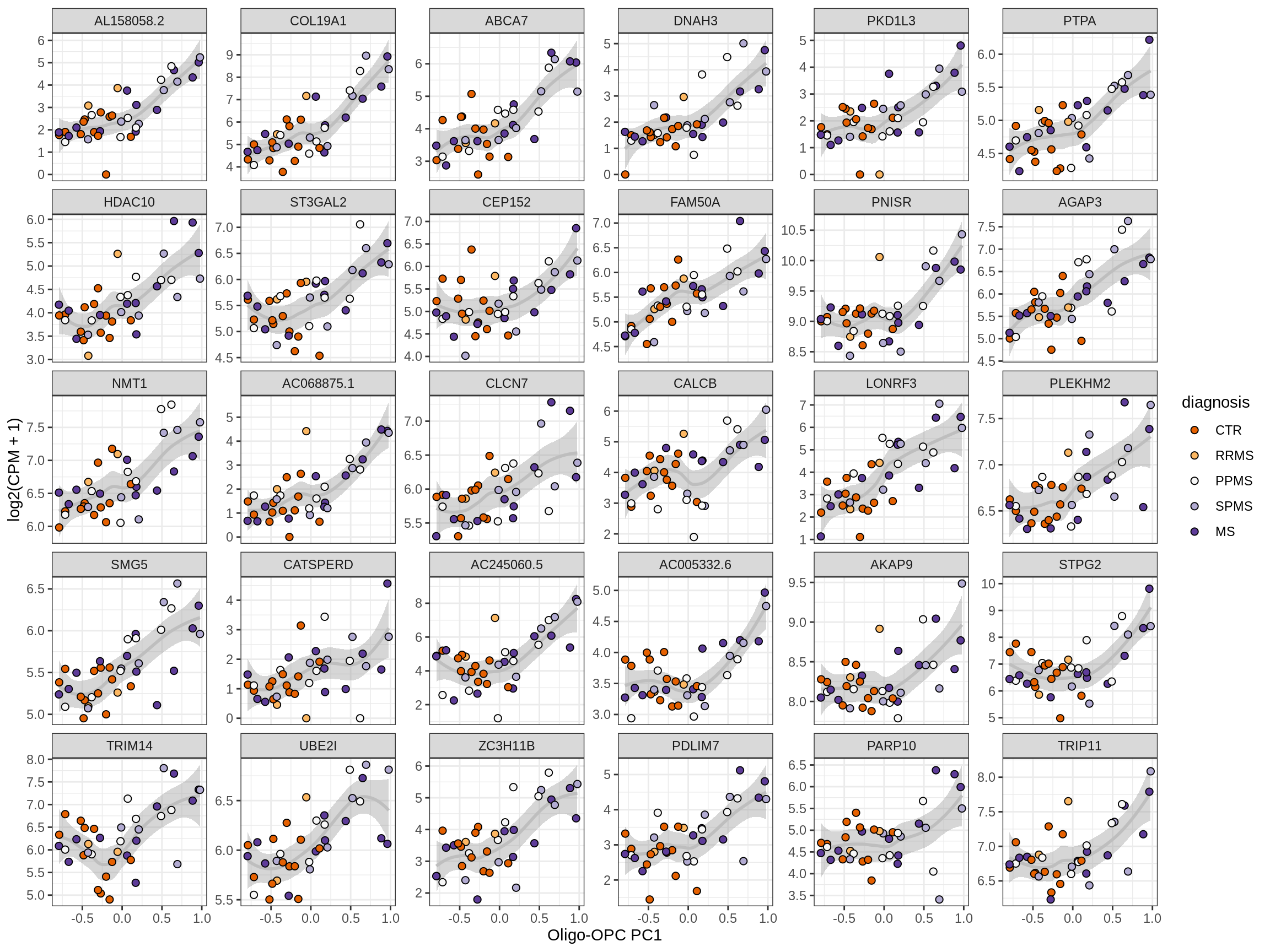
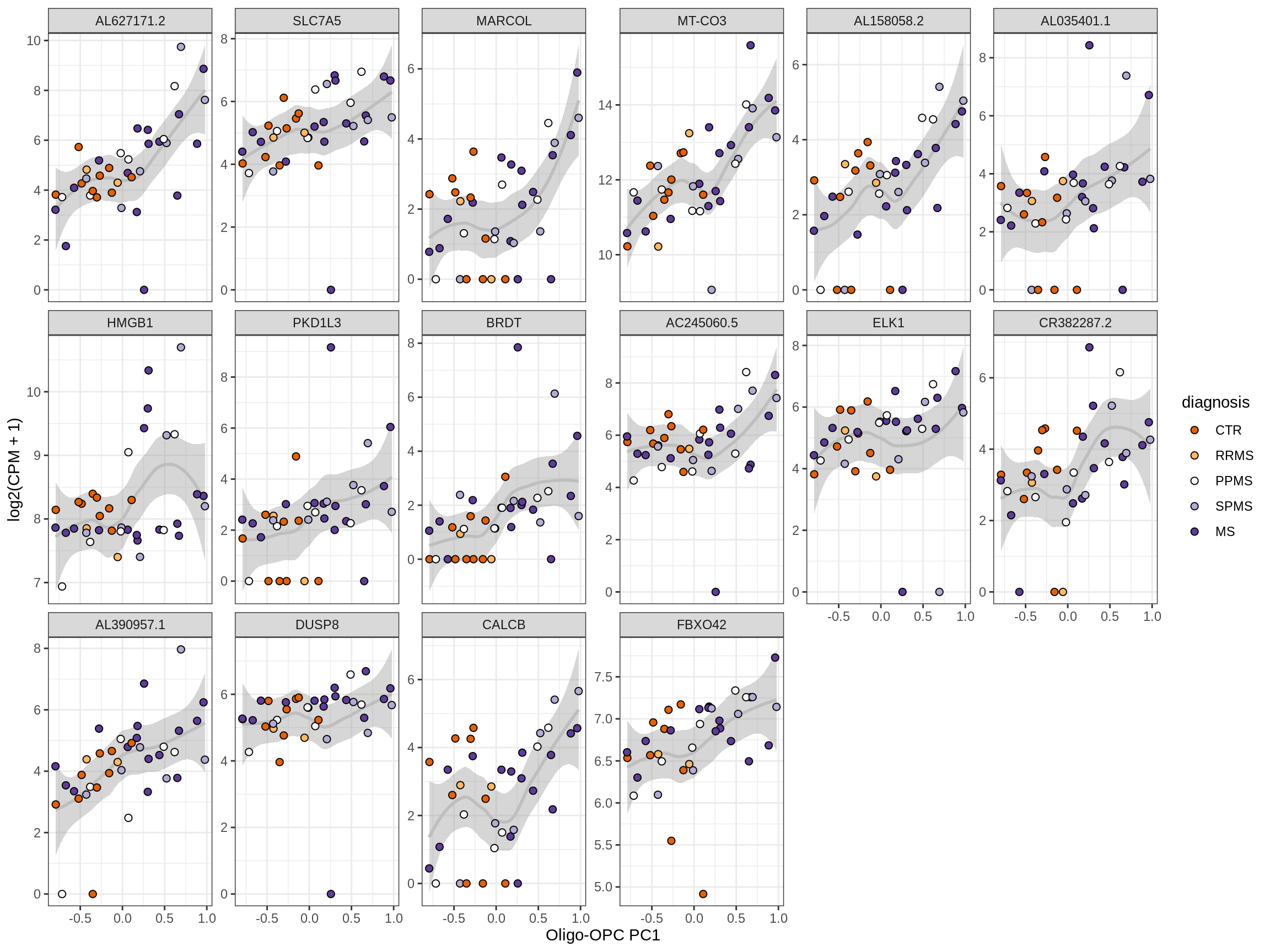
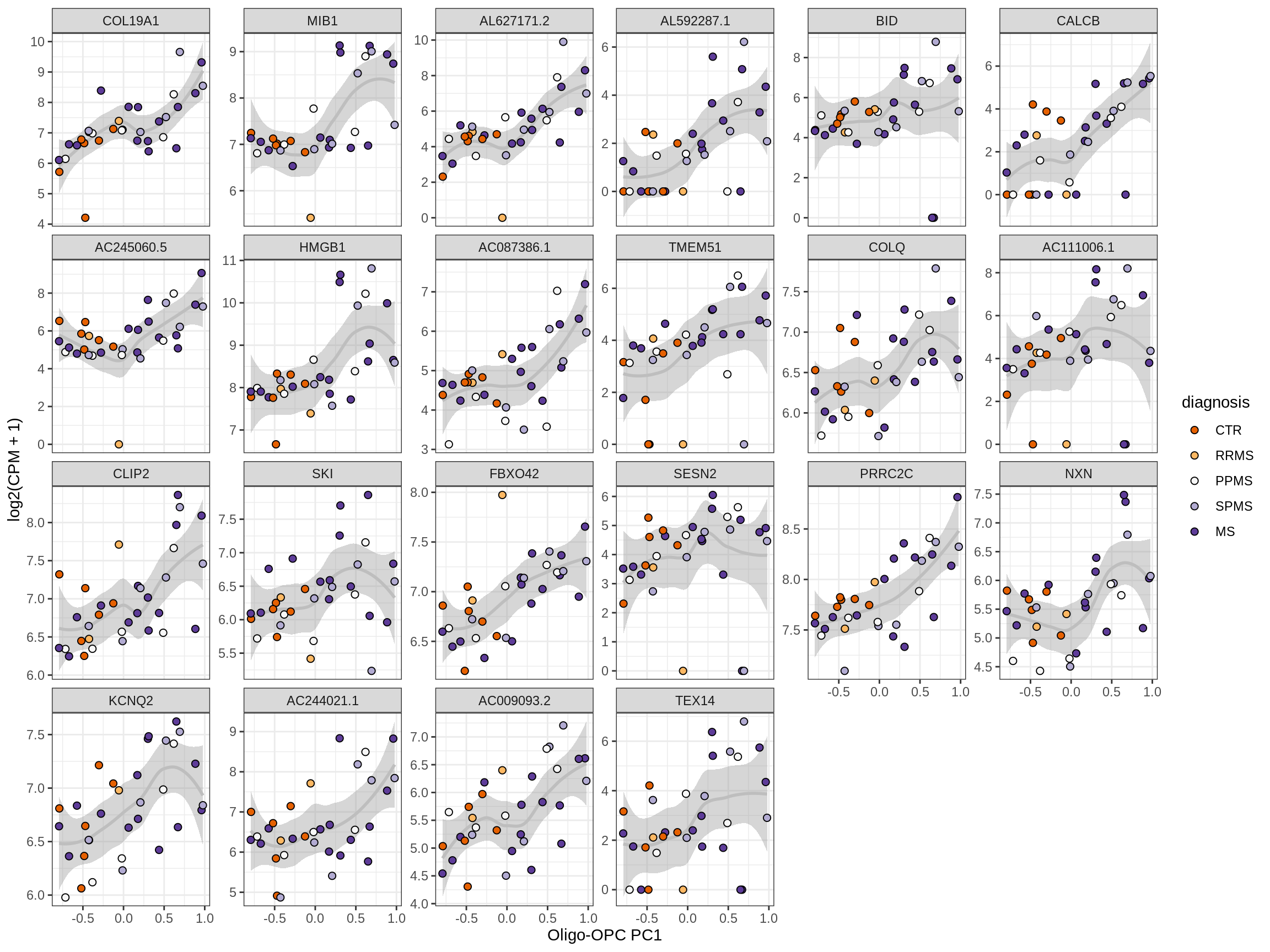
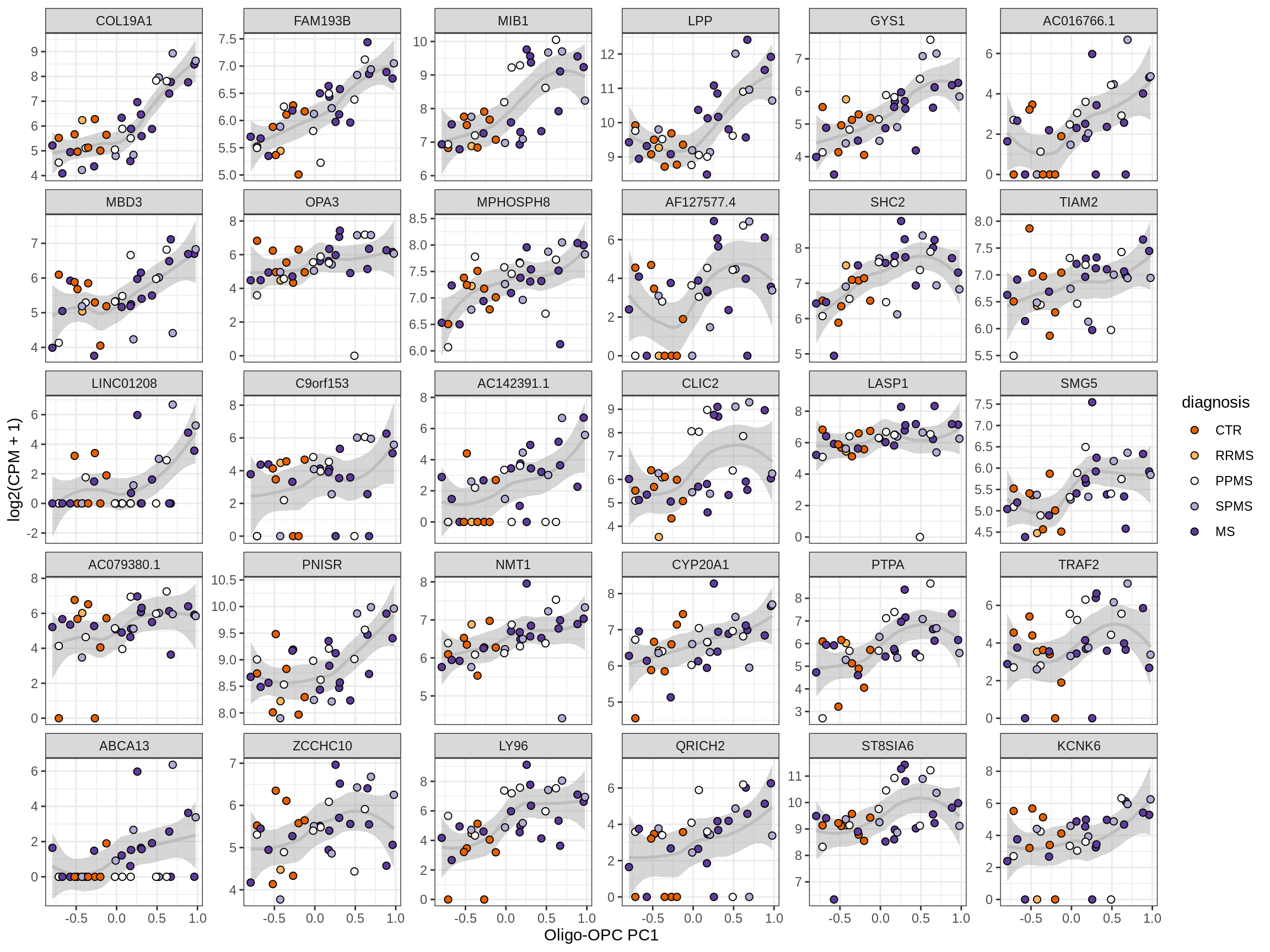
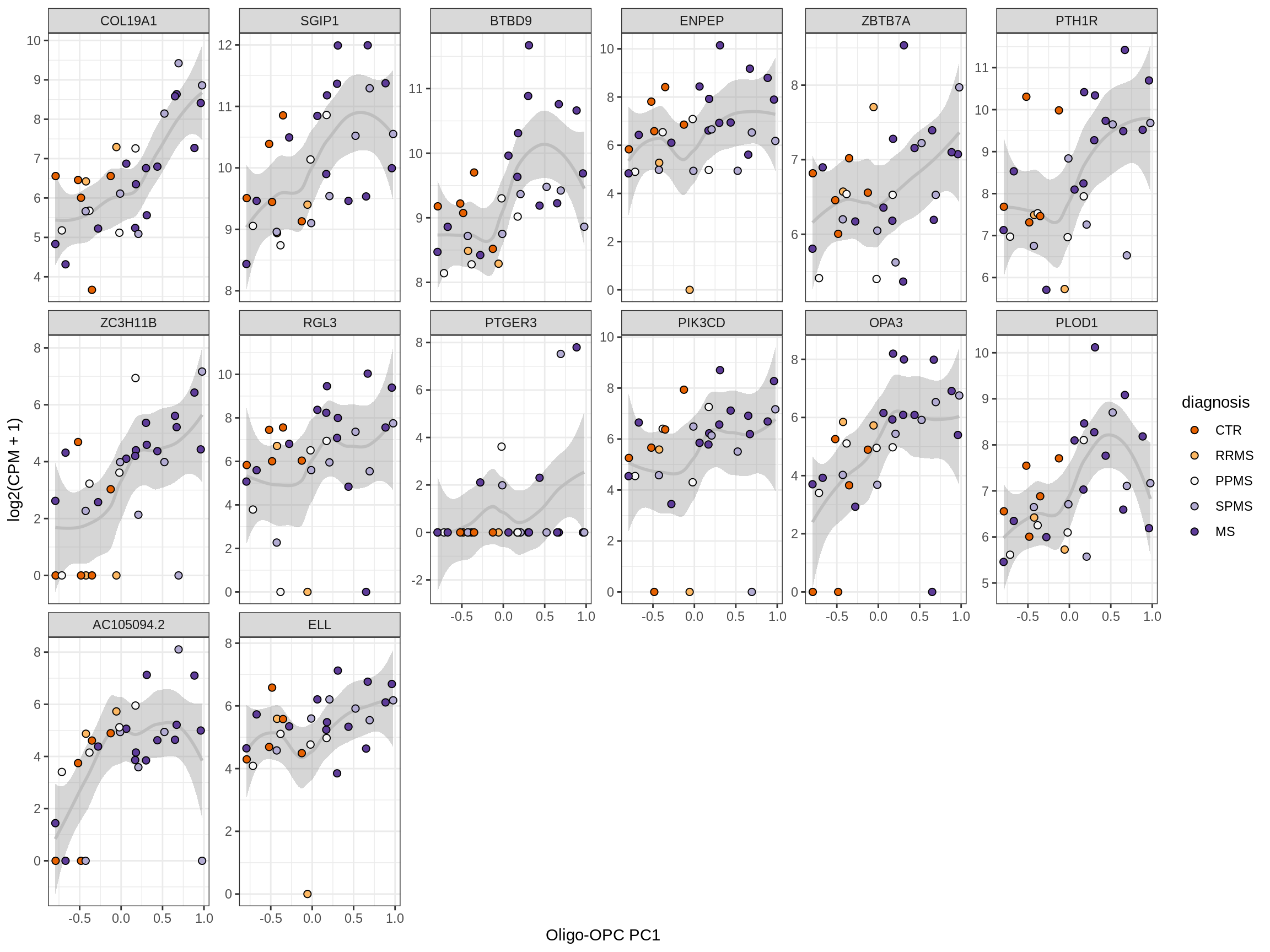
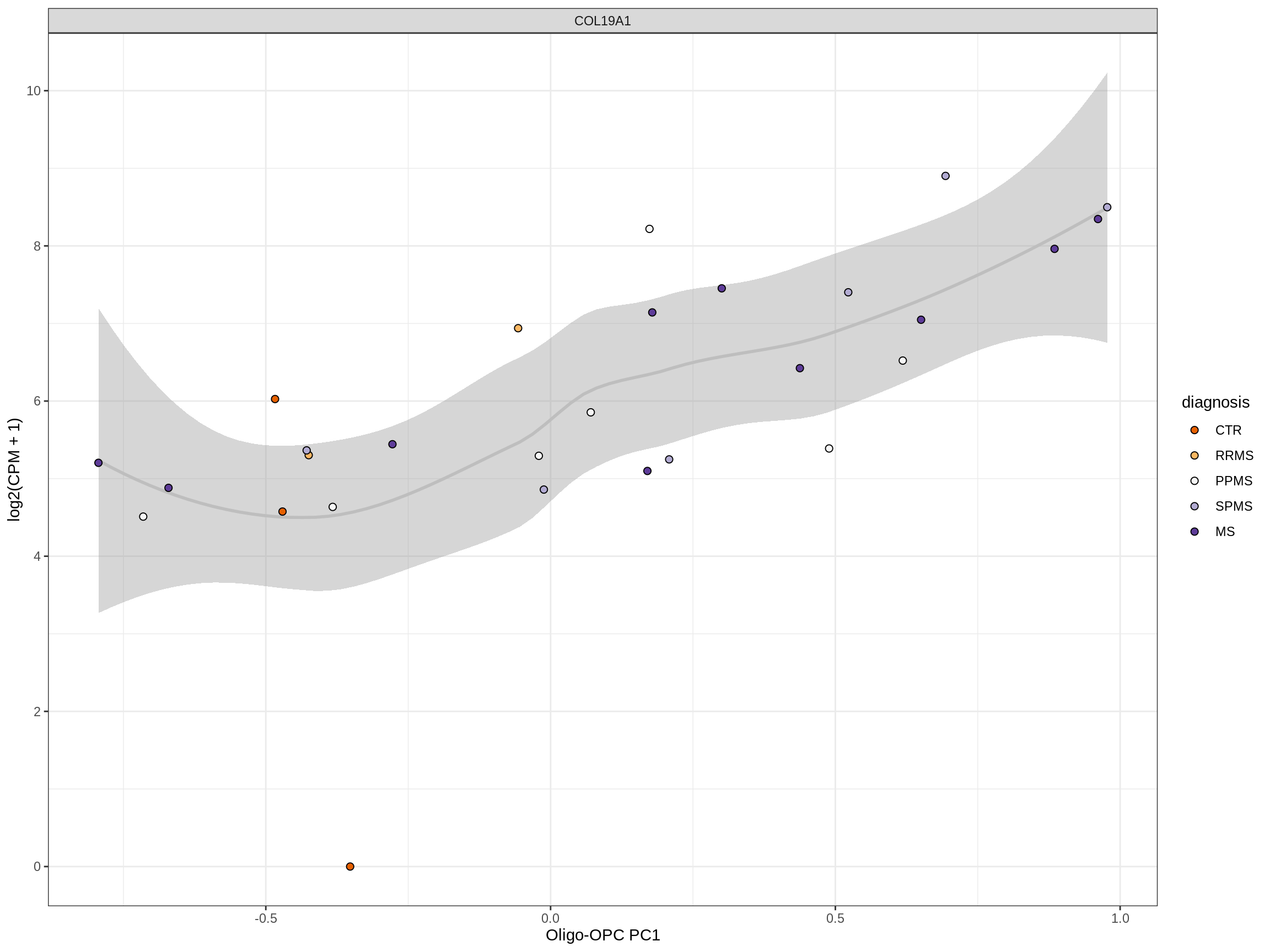
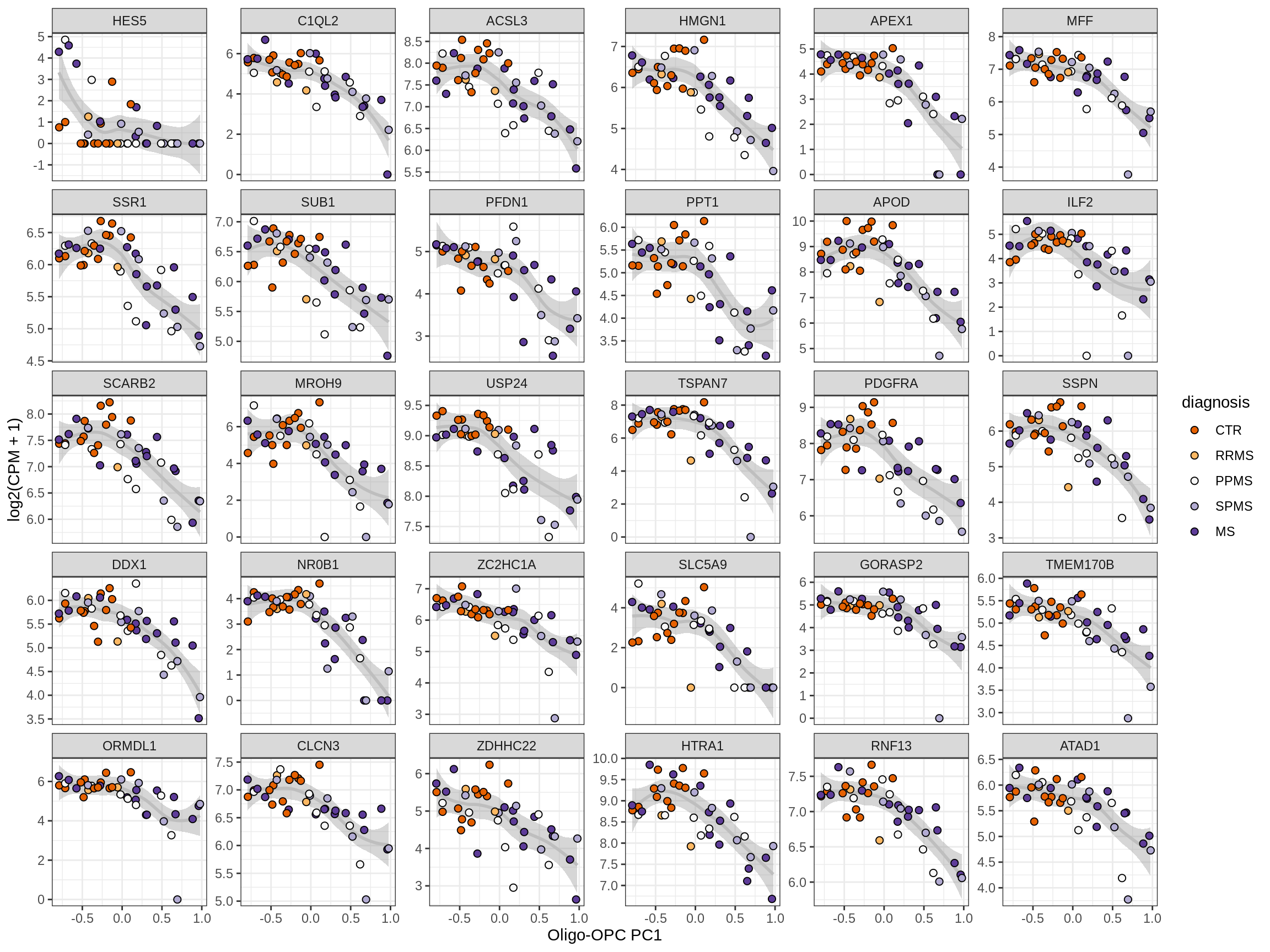
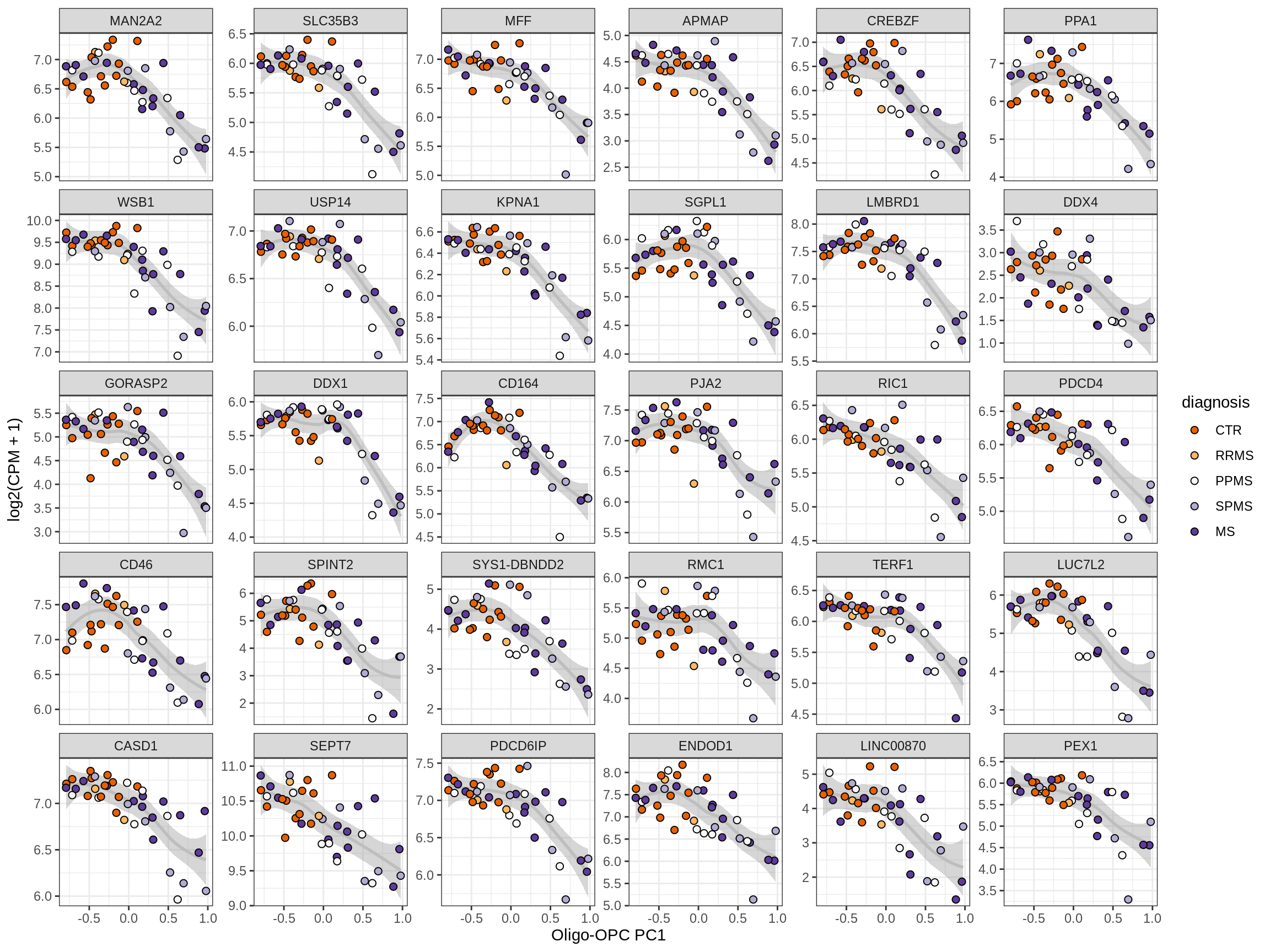
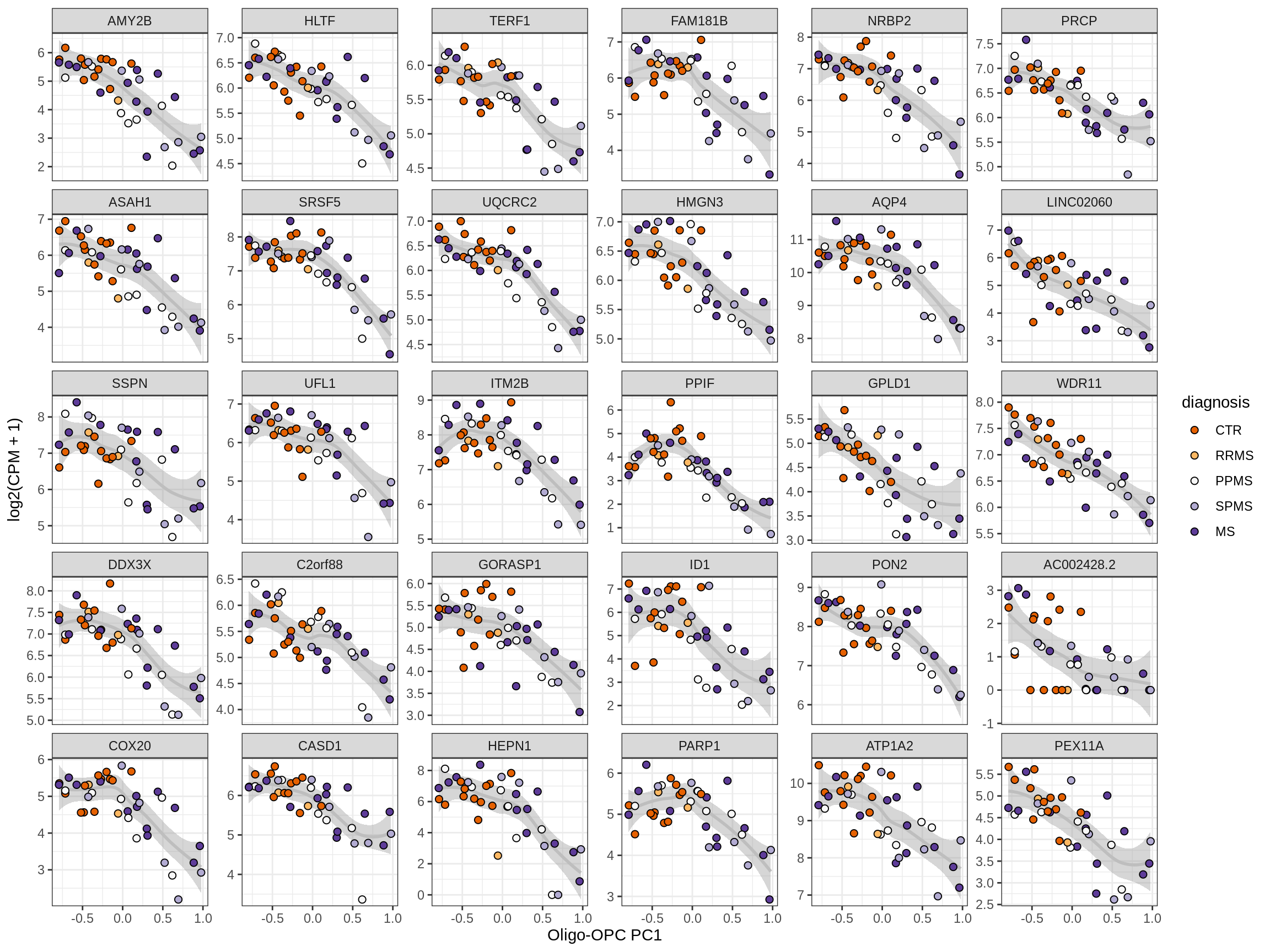
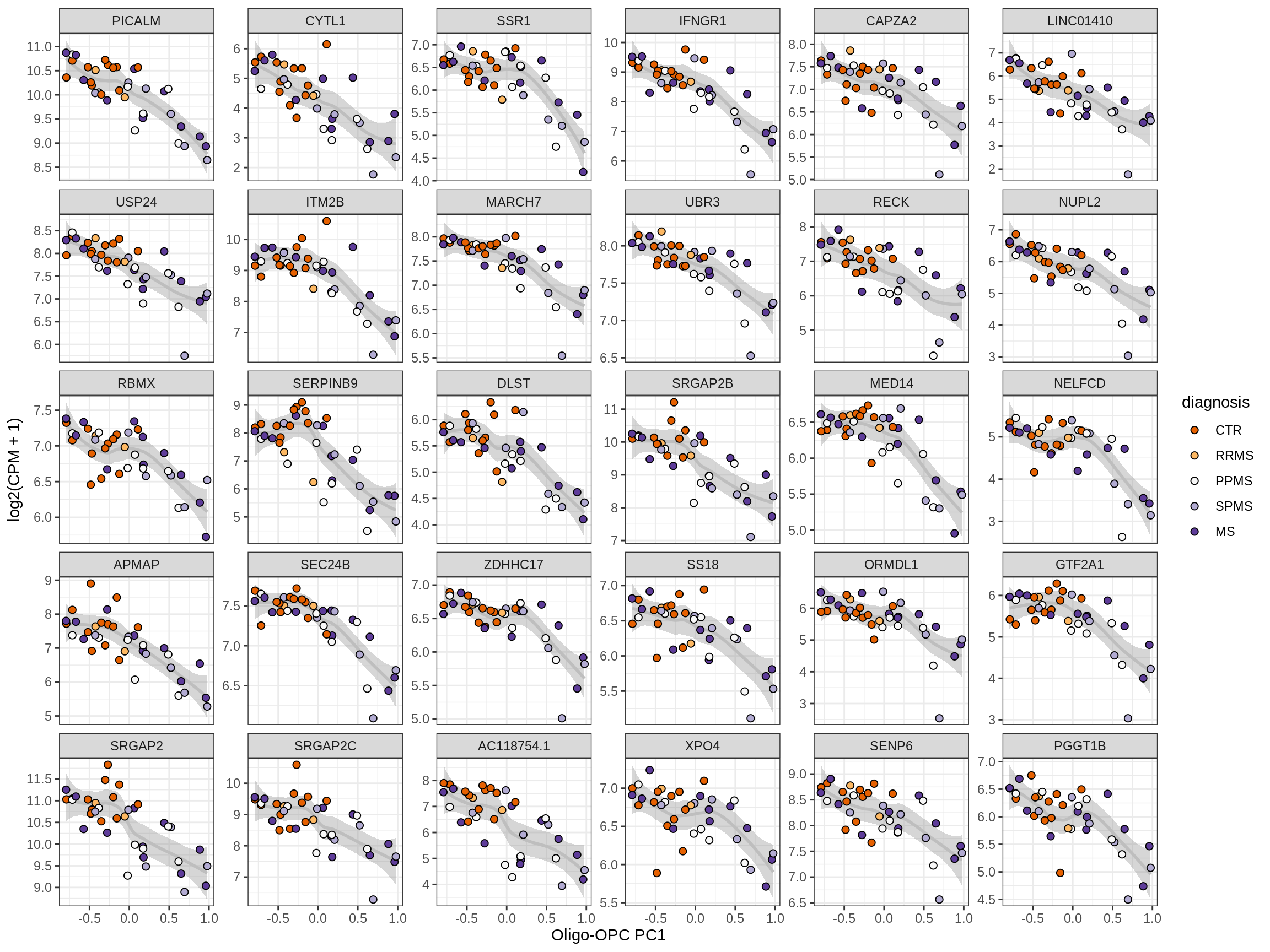
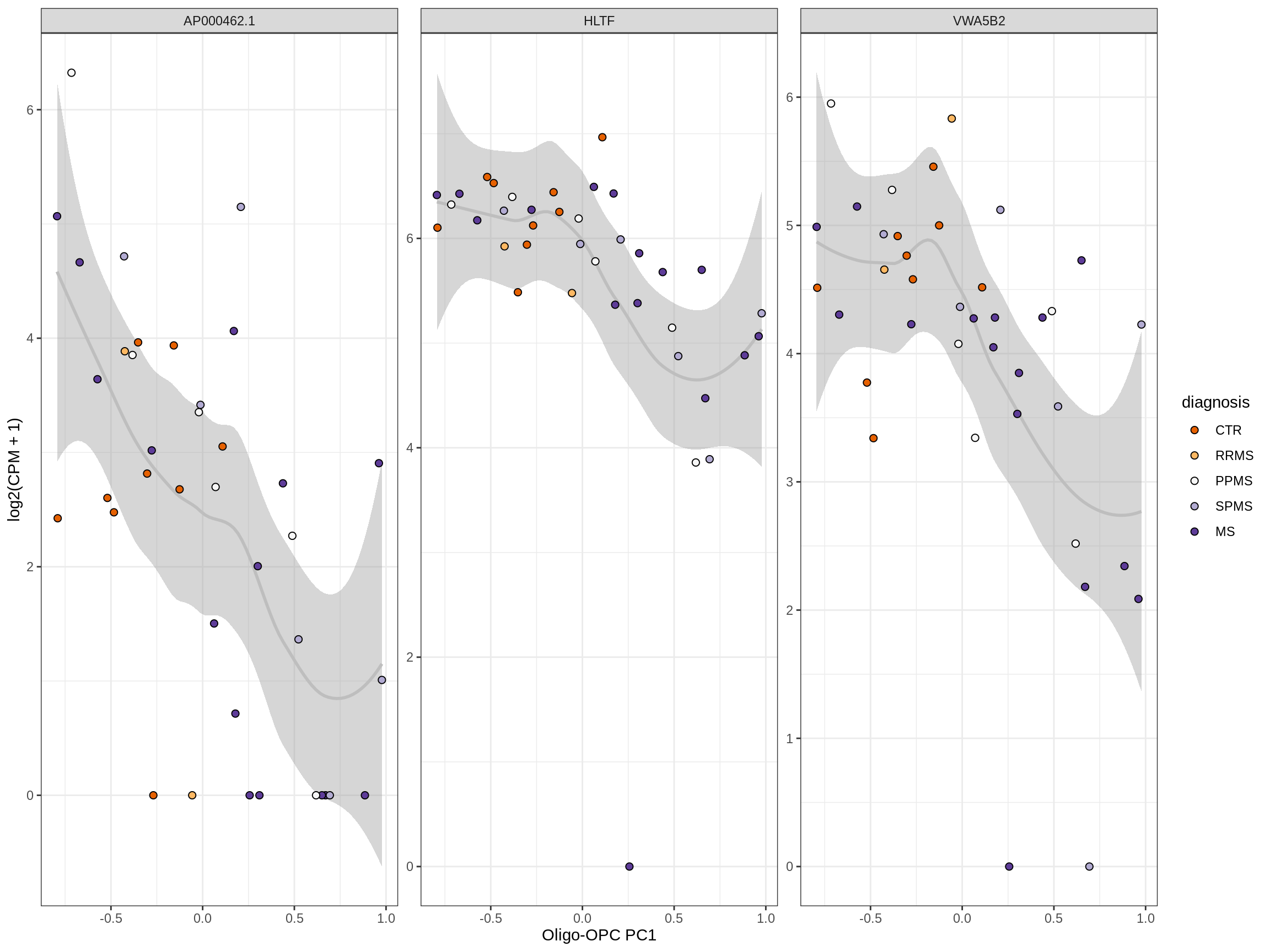
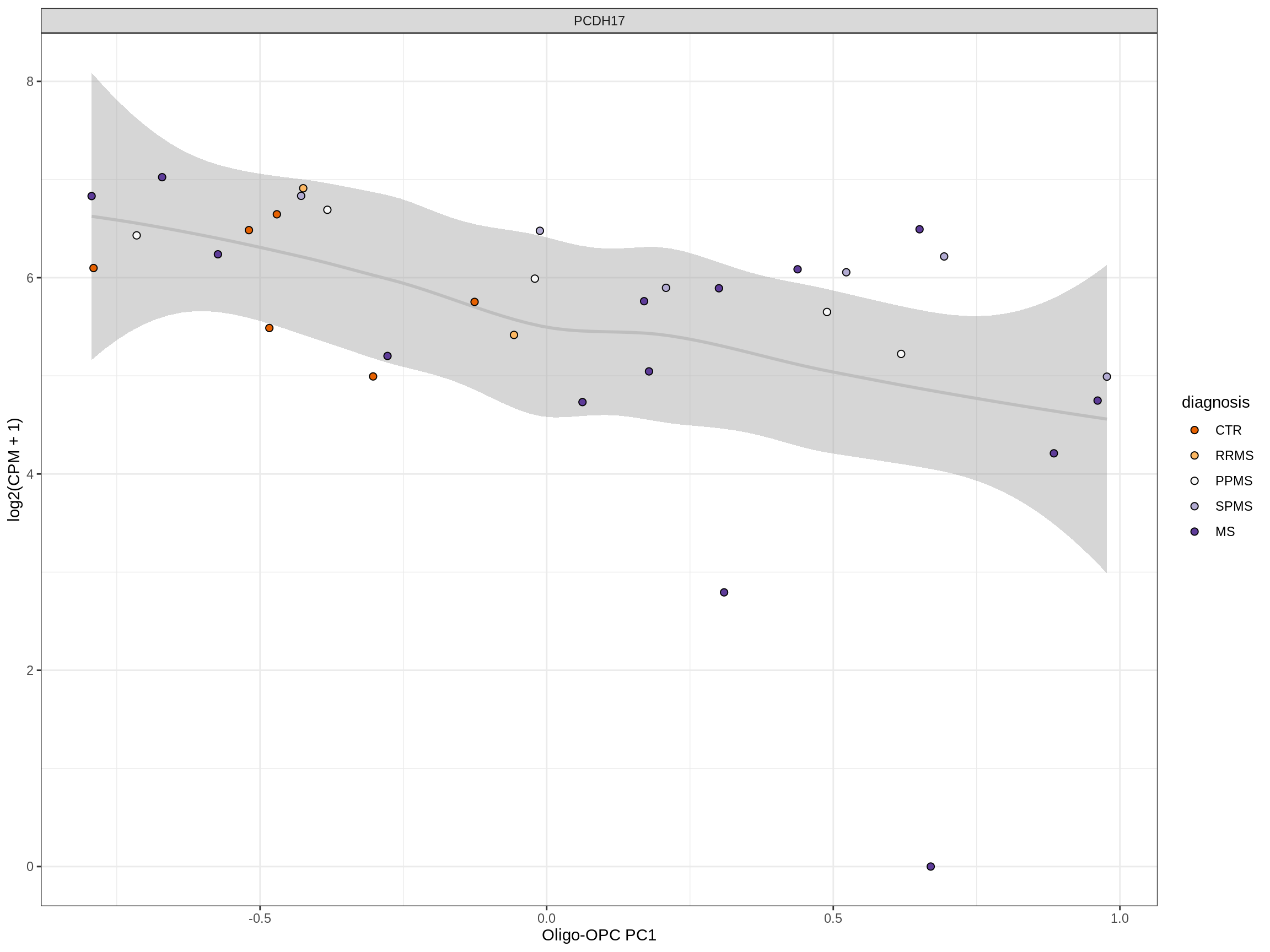
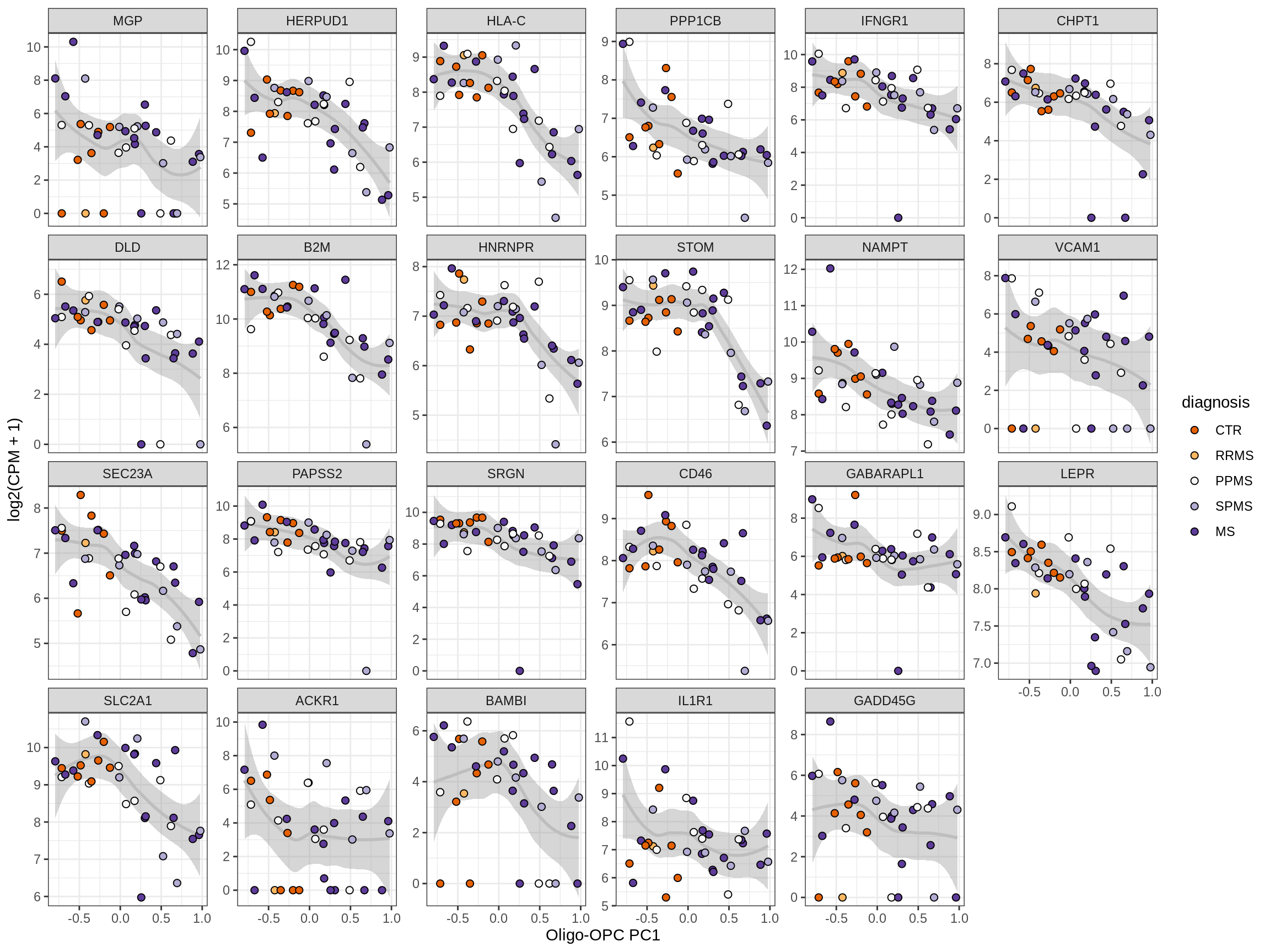
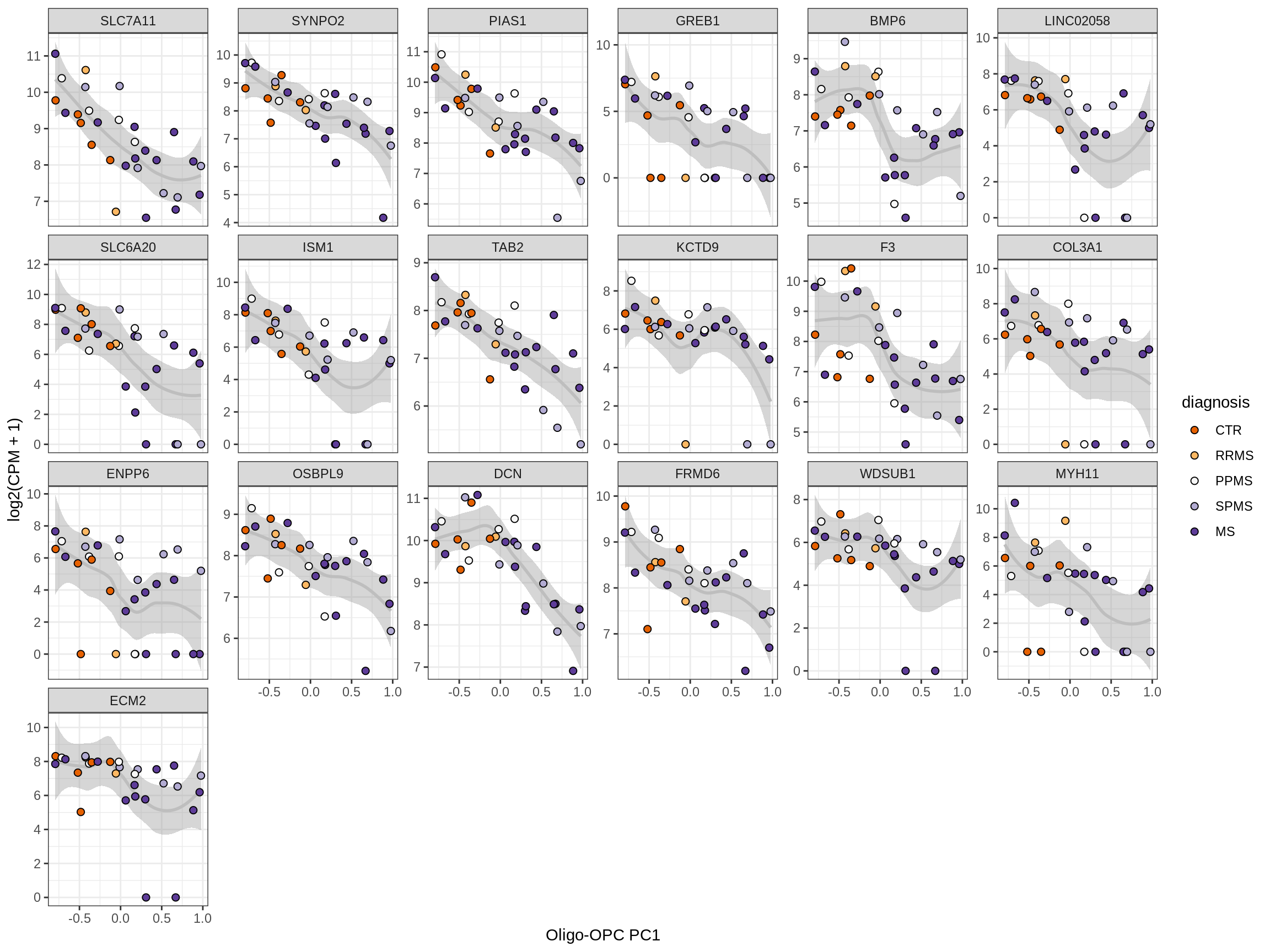
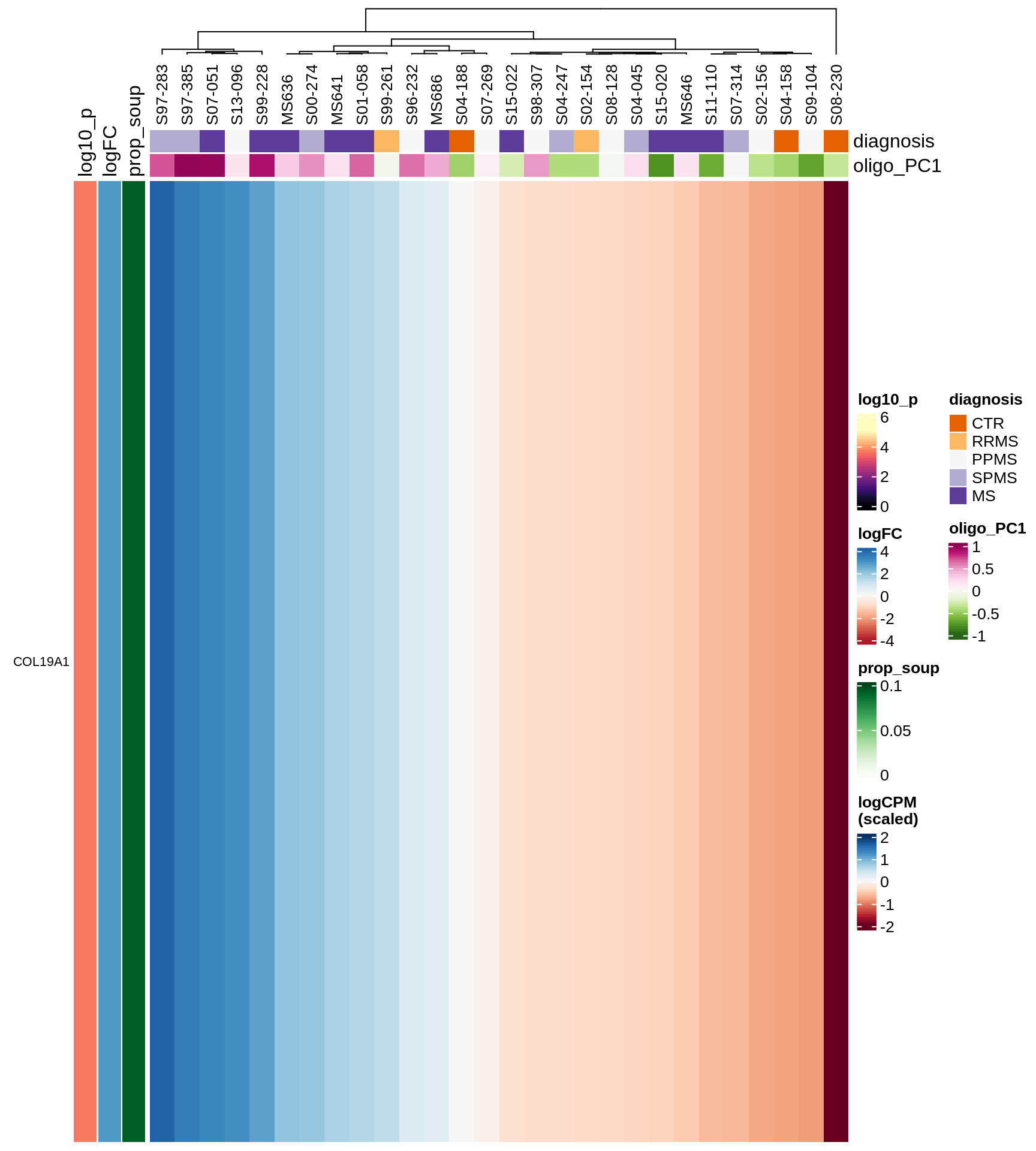
Heatmap of top oligo-PC1 genes
for (cl in broad_ord) {
# restrict to relevant GO terms
cat('## ', cl, '\n', sep='')
suppressWarnings(print(plot_oligo_pc_heatmap(cl, signif_dt,
cols_dt, pb_olg_pc, this_var, top_n = 60)))
cat('\n\n')
}OPCs / COPs
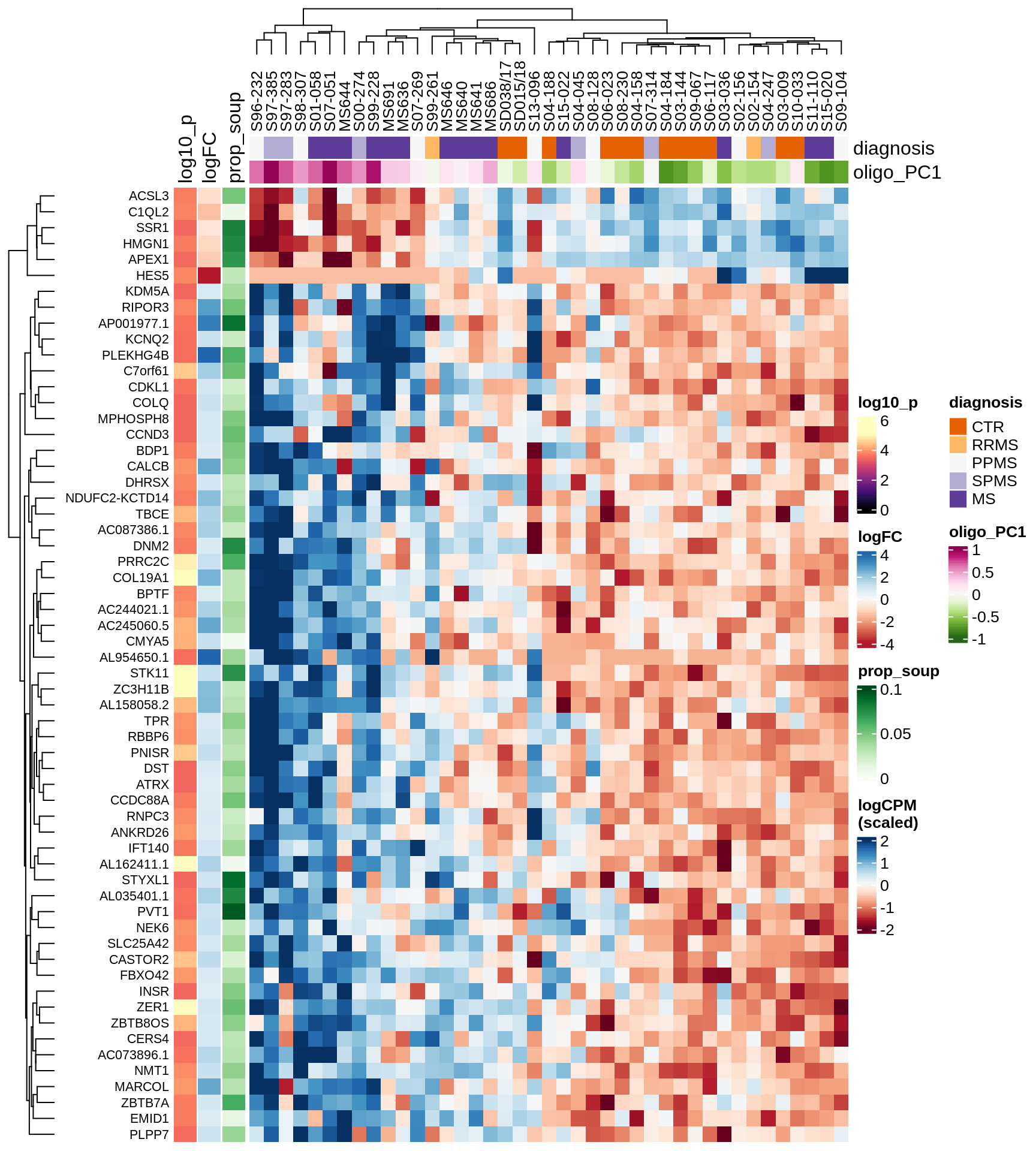
Oligodendrocytes
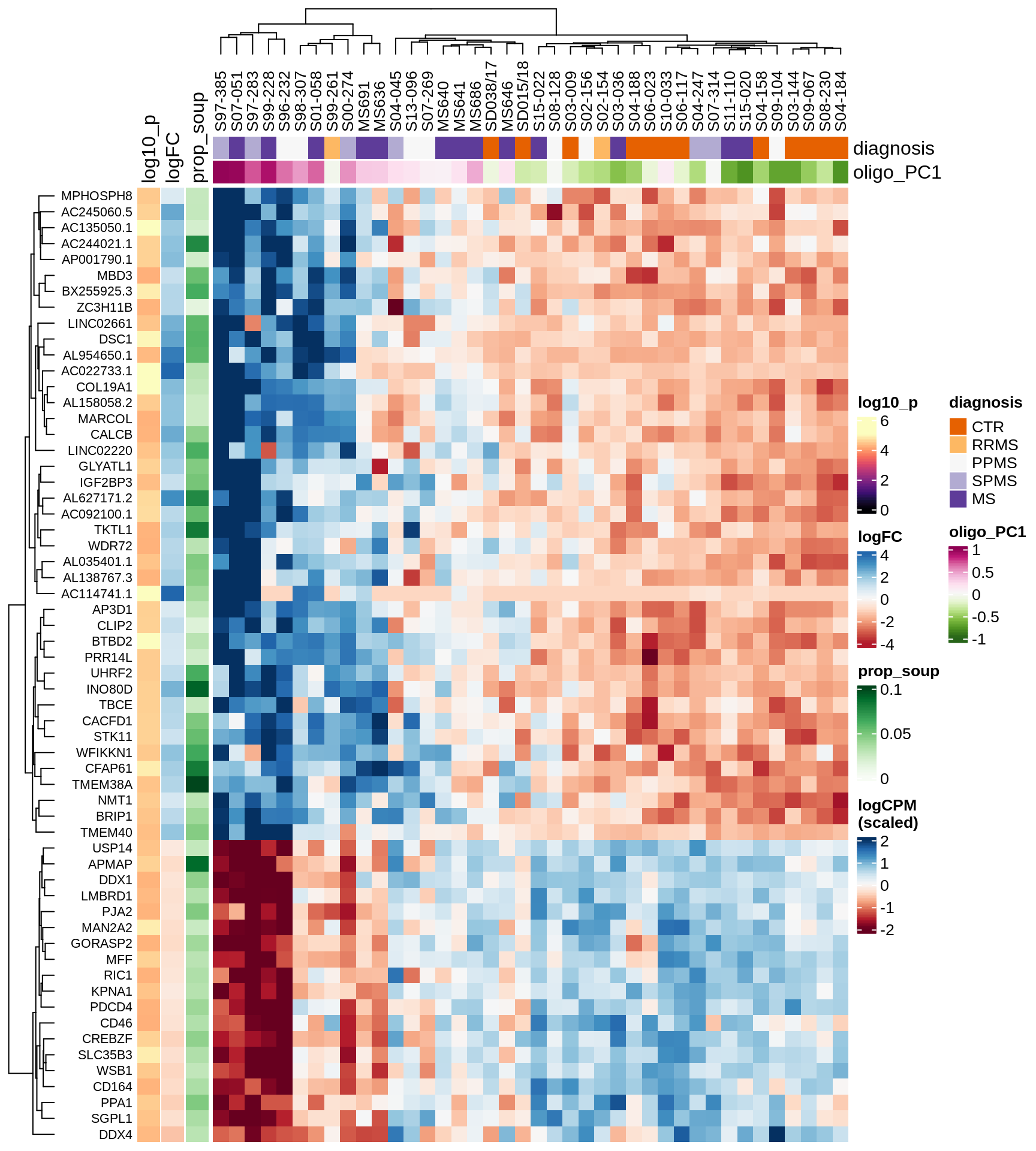
Astrocytes
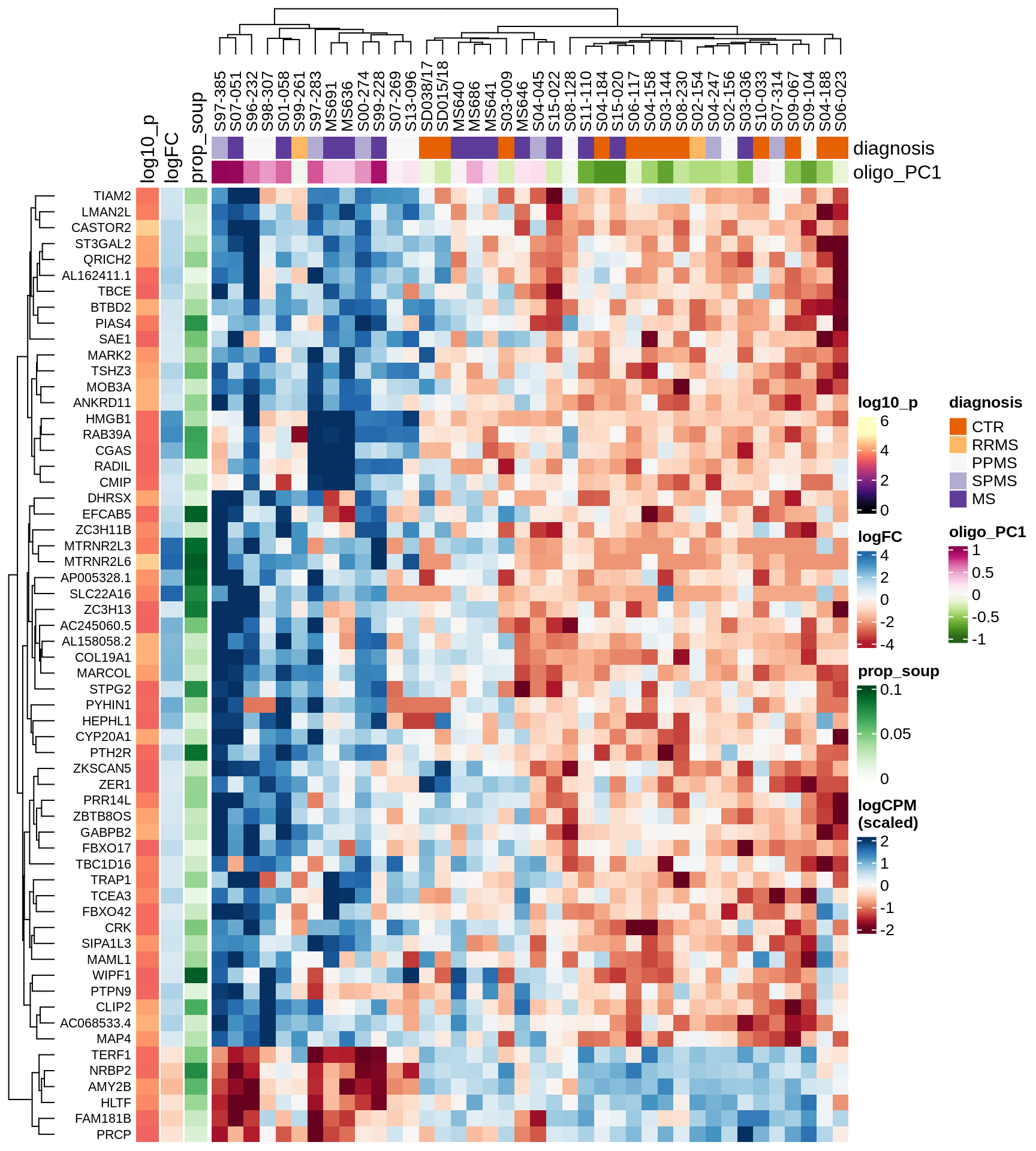
Microglia
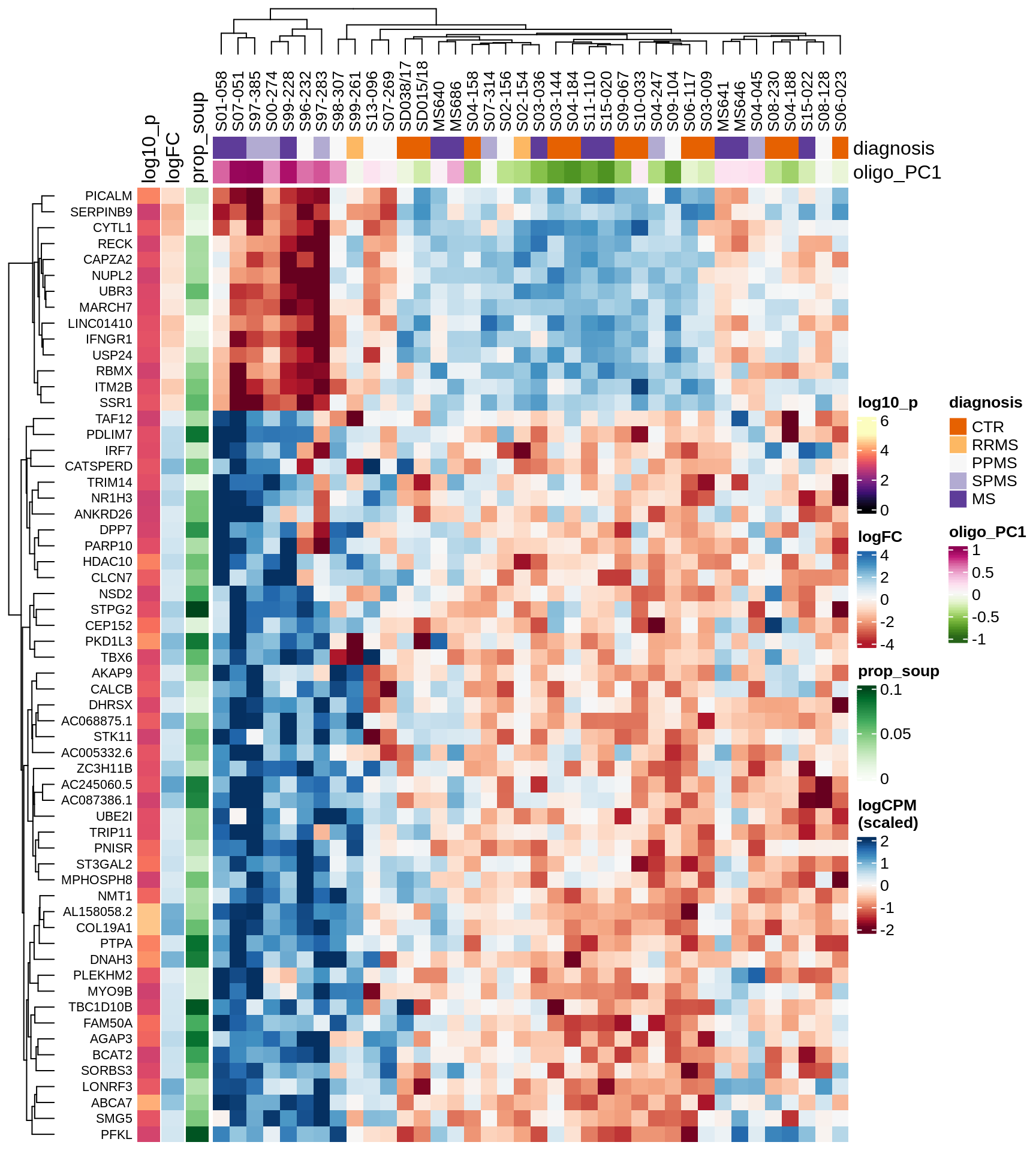
Excitatory neurons
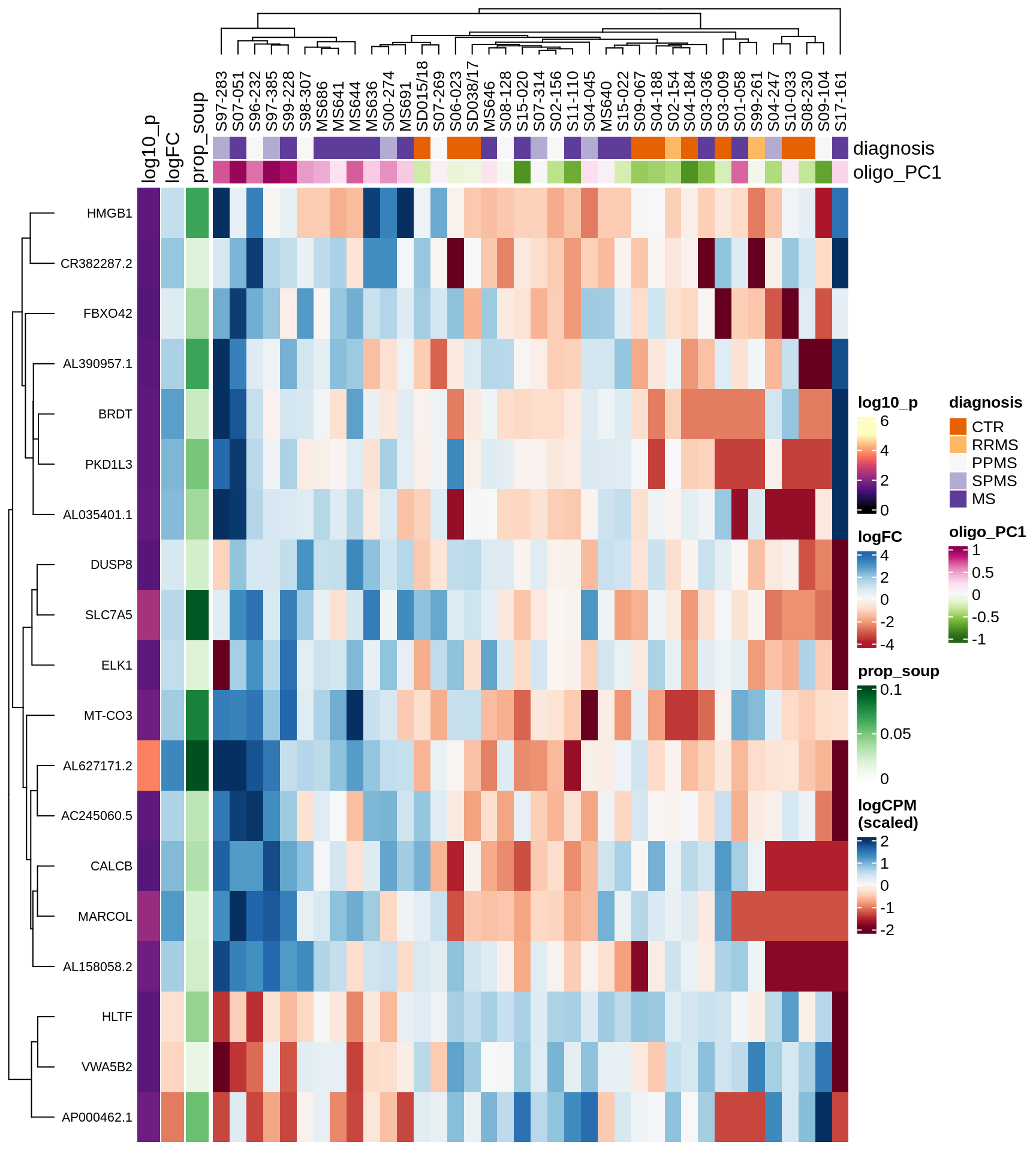
Inhibitory neurons
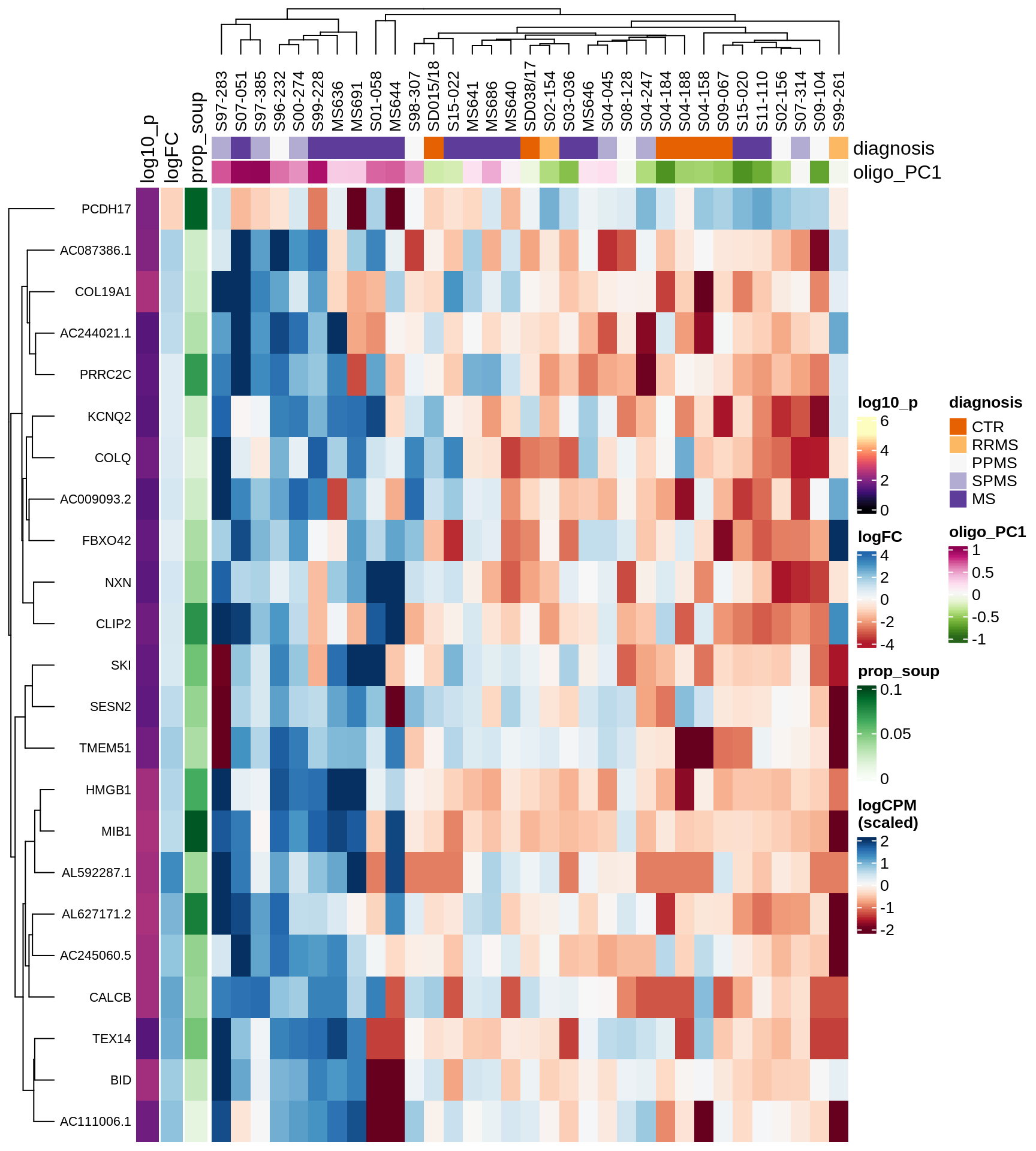
Endothelial cells
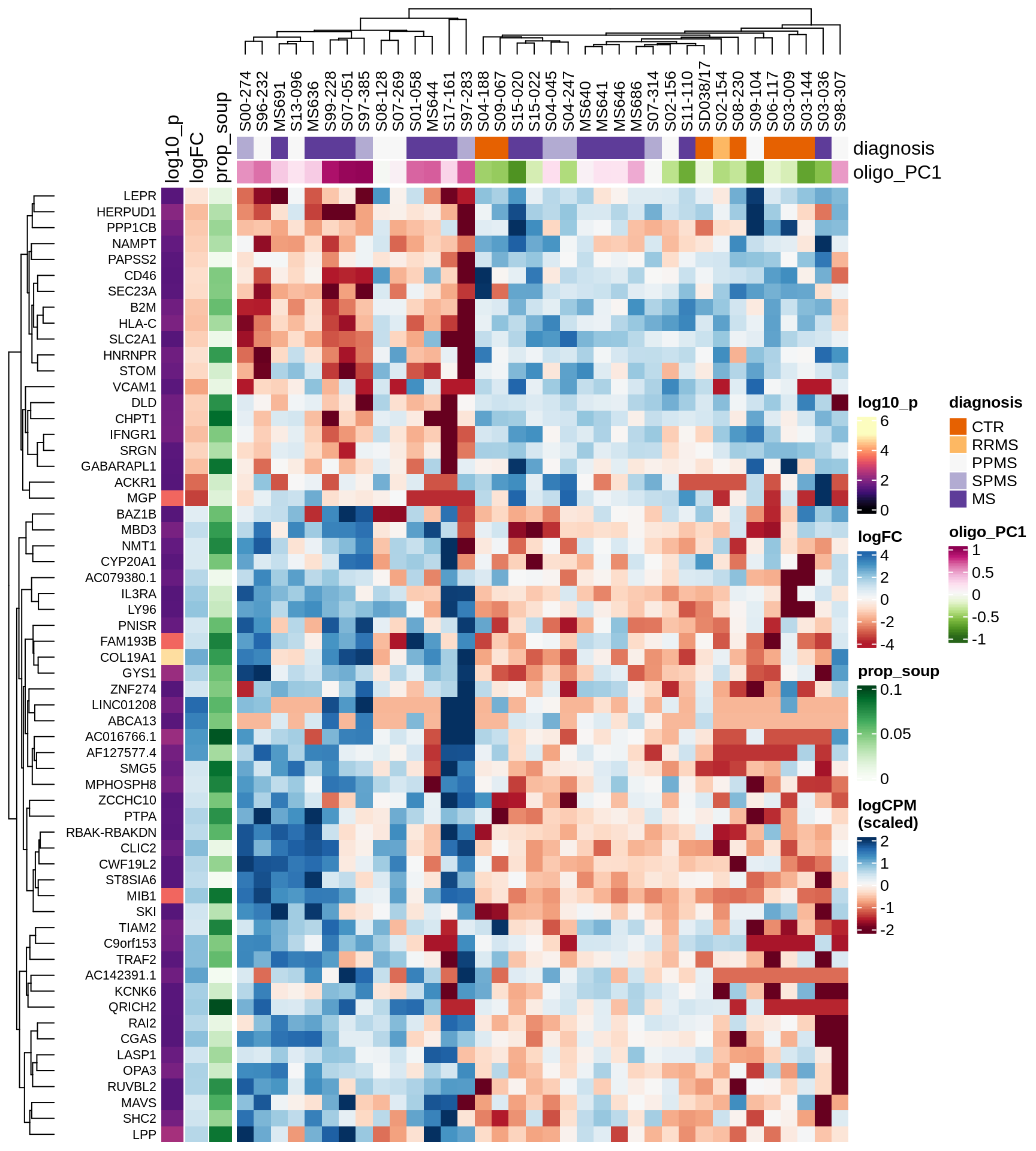
Pericytes
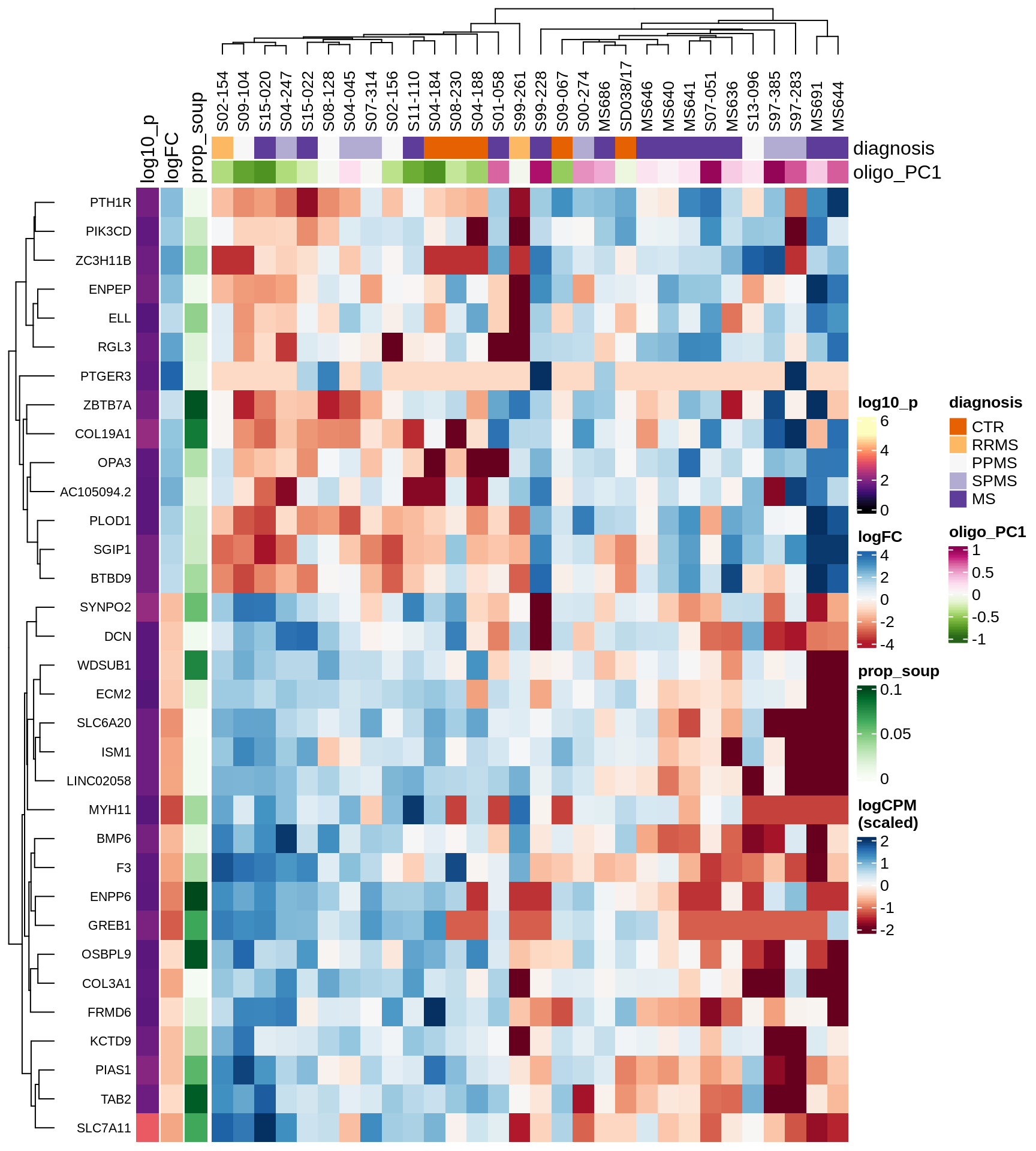
Immune
Could this be contamination?
Soup proportions vs logFCs
plot_dt = res_dt[ coef == 'oligo_pc1',
.(cluster_id, gene_id, symbol, soup_pc = 100 * mean_soup,
abs_logFC = abs(logFC), log_p = log10(p_adj.soup))] %>%
melt.data.table(measure = c('abs_logFC', 'log_p'),
variable.name = 'stat', value.name = 'value')
g = ggplot(plot_dt) +
aes(y = value, x = soup_pc) +
geom_bin2d() +
geom_smooth(method = 'gam', formula = y ~ s(x, bs = "cs"),
se = FALSE, colour = 'grey20') +
scale_x_continuous( breaks = pretty_breaks() ) +
scale_y_continuous( breaks = pretty_breaks() ) +
expand_limits( x = 0 ) +
scale_fill_distiller( palette = 'RdBu', trans = 'log10' ) +
facet_grid( stat ~ cluster_id, scales = 'free' ) +
theme_bw() + labs( x = '% soup', y = 'edgeR statistic' )
print(g)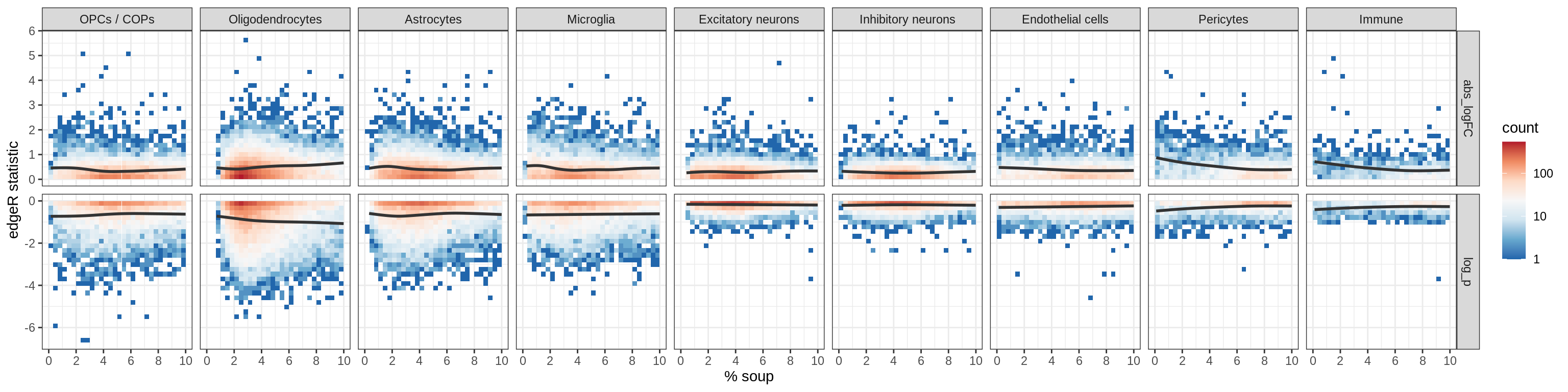
Oligo PCs vs QC stats
plot_dt = merge(qc_stats[str_detect(sample_id, 'WM')], olg_pc_dt,
by = 'patient_id')
g = ggplot(plot_dt) + aes(y = qc_val, x = oligo_pc1,
colour = type_broad, group = type_fine) +
geom_point(alpha = 0.1, size = 1) +
geom_smooth(se = FALSE, method = 'gam', formula = y ~ s(x, bs = 'cs')) +
scale_x_continuous( breaks = pretty_breaks() ) +
scale_y_continuous( breaks = pretty_breaks() ) +
scale_colour_manual( values = broad_cols ) +
facet_grid( qc_var ~ type_broad, scales = 'free_y' ) +
theme_bw() + theme(legend.position = 'bottom') +
labs( y = 'QC value', x = 'oligo PC1' )
(g)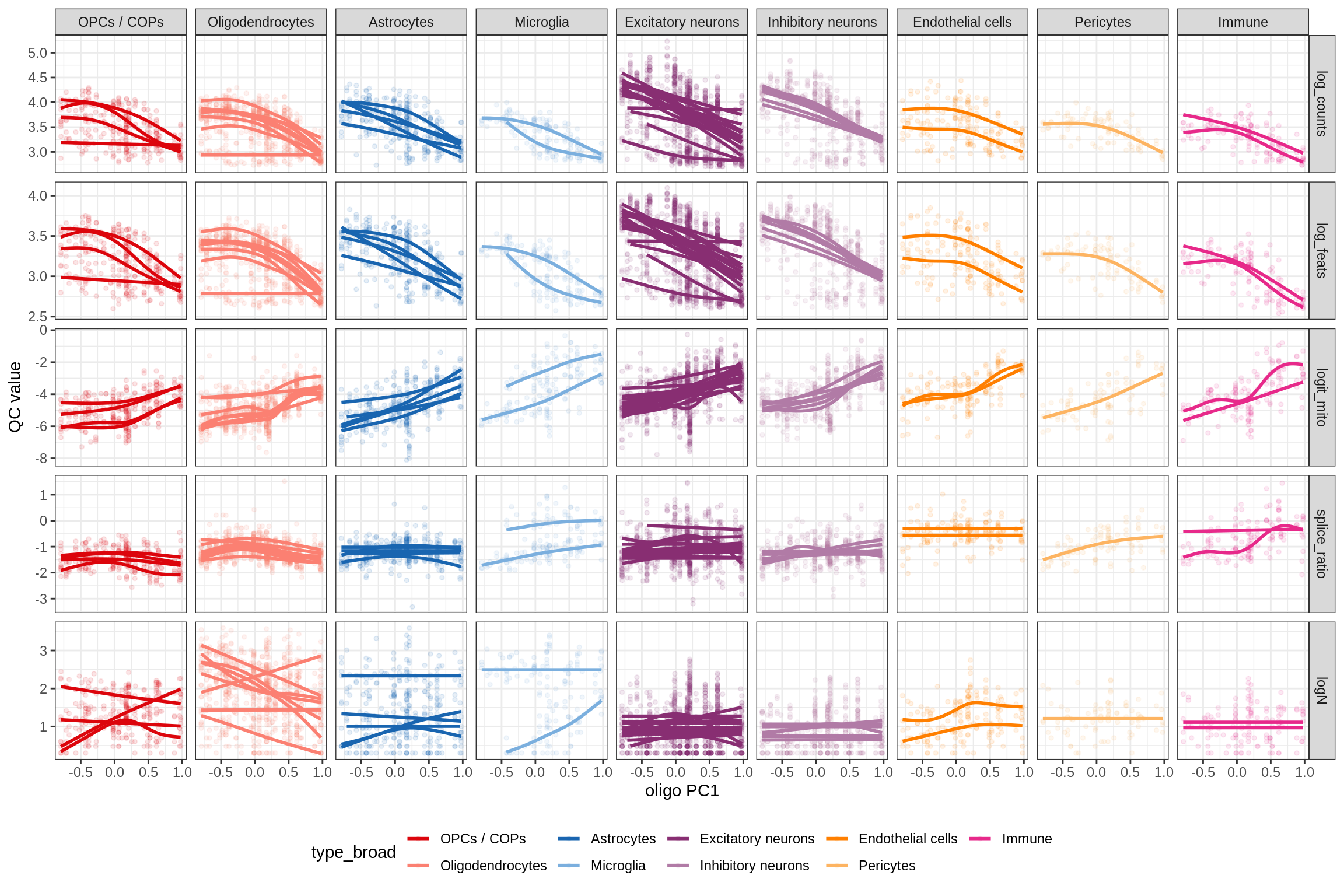
Outputs
devtools::session_info()- Session info ---------------------------------------------------------------
setting value
version R version 4.0.3 (2020-10-10)
os CentOS Linux 7 (Core)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_US.UTF-8
ctype C
tz Europe/Zurich
date 2021-07-01
- Packages -------------------------------------------------------------------
package * version date lib
ade4 1.7-16 2020-10-28 [1]
ANCOMBC * 1.0.5 2021-03-09 [1]
annotate 1.68.0 2020-10-27 [1]
AnnotationDbi 1.52.0 2020-10-27 [1]
ape 5.5 2021-04-25 [1]
assertthat * 0.2.1 2019-03-21 [2]
backports 1.2.1 2020-12-09 [2]
beachmat 2.6.4 2020-12-20 [1]
beeswarm 0.3.1 2021-03-07 [1]
Biobase * 2.50.0 2020-10-27 [1]
BiocGenerics * 0.36.1 2021-04-16 [1]
BiocManager 1.30.15 2021-05-11 [1]
BiocNeighbors 1.8.2 2020-12-07 [1]
BiocParallel * 1.24.1 2020-11-06 [1]
BiocSingular 1.6.0 2020-10-27 [1]
BiocStyle * 2.18.1 2020-11-24 [1]
biomformat 1.18.0 2020-10-27 [1]
Biostrings 2.58.0 2020-10-27 [1]
bit 4.0.4 2020-08-04 [2]
bit64 4.0.5 2020-08-30 [2]
bitops 1.0-7 2021-04-24 [2]
blme 1.0-5 2021-01-05 [1]
blob 1.2.1 2020-01-20 [2]
bluster 1.0.0 2020-10-27 [1]
boot 1.3-28 2021-05-03 [2]
broom 0.7.6 2021-04-05 [2]
bslib 0.2.5 2021-05-12 [2]
cachem 1.0.5 2021-05-15 [1]
Cairo 1.5-12.2 2020-07-07 [2]
callr 3.7.0 2021-04-20 [2]
caTools 1.18.2 2021-03-28 [2]
cellranger 1.1.0 2016-07-27 [2]
circlize * 0.4.12 2021-01-08 [1]
cli 2.5.0 2021-04-26 [2]
clue 0.3-59 2021-04-16 [1]
cluster 2.1.2 2021-04-17 [2]
codetools 0.2-18 2020-11-04 [2]
colorout * 1.2-2 2021-04-15 [1]
colorRamps 2.3 2012-10-29 [1]
colorspace 2.0-1 2021-05-04 [2]
ComplexHeatmap * 2.6.2 2020-11-12 [1]
crayon 1.4.1 2021-02-08 [2]
data.table * 1.14.0 2021-02-21 [2]
DBI 1.1.1 2021-01-15 [2]
dbplyr 2.1.1 2021-04-06 [2]
DelayedArray 0.16.3 2021-03-24 [1]
DelayedMatrixStats 1.12.3 2021-02-03 [1]
desc 1.3.0 2021-03-05 [2]
DESeq2 1.30.1 2021-02-19 [1]
devtools 2.4.1 2021-05-05 [1]
digest 0.6.27 2020-10-24 [2]
doParallel 1.0.16 2020-10-16 [1]
dplyr * 1.0.6 2021-05-05 [2]
dqrng 0.3.0 2021-05-01 [2]
DropletUtils * 1.10.3 2021-02-02 [1]
edgeR * 3.32.1 2021-01-14 [1]
ellipsis 0.3.2 2021-04-29 [2]
evaluate 0.14 2019-05-28 [2]
fansi 0.4.2 2021-01-15 [2]
farver 2.1.0 2021-02-28 [2]
fastmap 1.1.0 2021-01-25 [2]
fastmatch 1.1-0 2017-01-28 [1]
fgsea * 1.16.0 2020-10-27 [1]
forcats * 0.5.1 2021-01-27 [2]
foreach 1.5.1 2020-10-15 [2]
fs 1.5.0 2020-07-31 [2]
future 1.21.0 2020-12-10 [2]
future.apply 1.7.0 2021-01-04 [2]
genefilter 1.72.1 2021-01-21 [1]
geneplotter 1.68.0 2020-10-27 [1]
generics 0.1.0 2020-10-31 [2]
GenomeInfoDb * 1.26.7 2021-04-08 [1]
GenomeInfoDbData 1.2.4 2021-04-15 [1]
GenomicAlignments 1.26.0 2020-10-27 [1]
GenomicRanges * 1.42.0 2020-10-27 [1]
GetoptLong 1.0.5 2020-12-15 [1]
ggbeeswarm 0.6.0 2017-08-07 [1]
ggplot2 * 3.3.3 2020-12-30 [2]
ggrepel * 0.9.1 2021-01-15 [2]
git2r 0.28.0 2021-01-10 [1]
glmmTMB 1.0.2.1 2020-07-02 [1]
GlobalOptions 0.1.2 2020-06-10 [1]
globals 0.14.0 2020-11-22 [2]
glue 1.4.2 2020-08-27 [2]
googlesheets * 0.3.0 2018-06-29 [1]
gplots 3.1.1 2020-11-28 [2]
gridExtra 2.3 2017-09-09 [2]
grr 0.9.5 2016-08-26 [1]
gtable 0.3.0 2019-03-25 [2]
gtools 3.8.2 2020-03-31 [2]
haven 2.4.1 2021-04-23 [2]
HDF5Array 1.18.1 2021-02-04 [1]
highr 0.9 2021-04-16 [2]
hms 1.1.0 2021-05-17 [2]
htmltools 0.5.1.1 2021-01-22 [2]
httpuv 1.6.1 2021-05-07 [2]
httr 1.4.2 2020-07-20 [2]
igraph 1.2.6 2020-10-06 [2]
IRanges * 2.24.1 2020-12-12 [1]
irlba 2.3.3 2019-02-05 [2]
iterators 1.0.13 2020-10-15 [2]
janitor 2.1.0 2021-01-05 [1]
jquerylib 0.1.4 2021-04-26 [2]
jsonlite 1.7.2 2020-12-09 [2]
KernSmooth 2.23-20 2021-05-03 [2]
knitr 1.33 2021-04-24 [1]
labeling 0.4.2 2020-10-20 [2]
later 1.2.0 2021-04-23 [2]
lattice 0.20-44 2021-05-02 [2]
lifecycle 1.0.0 2021-02-15 [2]
limma * 3.46.0 2020-10-27 [1]
listenv 0.8.0 2019-12-05 [2]
lme4 1.1-27 2021-05-15 [1]
lmerTest 3.1-3 2020-10-23 [1]
locfit 1.5-9.4 2020-03-25 [1]
lubridate 1.7.10 2021-02-26 [2]
magrittr * 2.0.1 2020-11-17 [1]
MASS * 7.3-54 2021-05-03 [2]
Matrix * 1.3-3 2021-05-04 [2]
Matrix.utils * 0.9.8 2020-02-26 [1]
MatrixGenerics * 1.2.1 2021-01-30 [1]
matrixStats * 0.59.0 2021-06-01 [1]
memoise 2.0.0 2021-01-26 [1]
mgcv 1.8-35 2021-04-18 [2]
microbiome 1.12.0 2020-10-27 [1]
minqa 1.2.4 2014-10-09 [1]
modelr 0.1.8 2020-05-19 [2]
multtest 2.46.0 2020-10-27 [1]
munsell 0.5.0 2018-06-12 [2]
muscat * 1.5.1 2021-04-15 [1]
nlme 3.1-152 2021-02-04 [2]
nloptr 1.2.2.2 2020-07-02 [1]
nnls * 1.4 2012-03-19 [1]
numDeriv 2016.8-1.1 2019-06-06 [2]
parallelly 1.25.0 2021-04-30 [2]
patchwork * 1.1.1 2020-12-17 [2]
pbkrtest 0.5.1 2021-03-09 [1]
permute 0.9-5 2019-03-12 [1]
phyloseq * 1.34.0 2020-10-27 [1]
pillar 1.6.1 2021-05-16 [2]
pkgbuild 1.2.0 2020-12-15 [1]
pkgconfig 2.0.3 2019-09-22 [2]
pkgload 1.2.1 2021-04-06 [2]
plyr 1.8.6 2020-03-03 [2]
png 0.1-7 2013-12-03 [2]
prettyunits 1.1.1 2020-01-24 [2]
processx 3.5.2 2021-04-30 [2]
progress 1.2.2 2019-05-16 [2]
promises 1.2.0.1 2021-02-11 [2]
ps 1.6.0 2021-02-28 [2]
purrr * 0.3.4 2020-04-17 [2]
R.methodsS3 1.8.1 2020-08-26 [1]
R.oo 1.24.0 2020-08-26 [1]
R.utils 2.10.1 2020-08-26 [1]
R6 2.5.0 2020-10-28 [2]
rbibutils 2.1.1 2021-04-28 [1]
RColorBrewer * 1.1-2 2014-12-07 [2]
Rcpp 1.0.6 2021-01-15 [2]
RCurl 1.98-1.3 2021-03-16 [1]
Rdpack 2.1.1 2021-02-23 [1]
readr * 1.4.0 2020-10-05 [2]
readxl * 1.3.1 2019-03-13 [2]
registry 0.5-1 2019-03-05 [1]
remotes 2.3.0 2021-04-01 [1]
reprex 2.0.0 2021-04-02 [2]
reshape2 * 1.4.4 2020-04-09 [2]
reticulate * 1.20 2021-05-03 [2]
rhdf5 2.34.0 2020-10-27 [1]
rhdf5filters 1.2.1 2021-05-03 [1]
Rhdf5lib 1.12.1 2021-01-26 [1]
rjson 0.2.20 2018-06-08 [1]
rlang 0.4.11 2021-04-30 [2]
rmarkdown * 2.8 2021-05-07 [2]
rprojroot 2.0.2 2020-11-15 [2]
Rsamtools 2.6.0 2020-10-27 [1]
RSQLite 2.2.7 2021-04-22 [1]
rstudioapi 0.13 2020-11-12 [2]
rsvd 1.0.5 2021-04-16 [1]
rtracklayer * 1.50.0 2020-10-27 [1]
Rtsne 0.15 2018-11-10 [2]
rvest 1.0.0 2021-03-09 [2]
S4Vectors * 0.28.1 2020-12-09 [1]
sass 0.4.0 2021-05-12 [2]
scales * 1.1.1 2020-05-11 [2]
scater * 1.18.6 2021-02-26 [1]
scran * 1.18.7 2021-04-16 [1]
sctransform 0.3.2 2020-12-16 [2]
scuttle 1.0.4 2020-12-17 [1]
seriation * 1.2-9 2020-10-01 [1]
sessioninfo 1.1.1 2018-11-05 [1]
shape 1.4.6 2021-05-19 [1]
SingleCellExperiment * 1.12.0 2020-10-27 [1]
snakecase 0.11.0 2019-05-25 [1]
sparseMatrixStats 1.2.1 2021-02-02 [1]
statmod 1.4.36 2021-05-10 [1]
stringi 1.6.2 2021-05-17 [2]
stringr * 1.4.0 2019-02-10 [2]
SummarizedExperiment * 1.20.0 2020-10-27 [1]
survival 3.2-11 2021-04-26 [2]
testthat 3.0.2 2021-02-14 [2]
tibble * 3.1.2 2021-05-16 [2]
tictoc * 1.0.1 2021-04-19 [1]
tidyr * 1.1.3 2021-03-03 [2]
tidyselect 1.1.1 2021-04-30 [2]
tidyverse * 1.3.1 2021-04-15 [2]
TMB 1.7.20 2021-04-08 [1]
TSP 1.1-10 2020-04-17 [1]
UpSetR * 1.4.0 2019-05-22 [1]
usethis 2.0.1 2021-02-10 [1]
utf8 1.2.1 2021-03-12 [2]
variancePartition 1.20.0 2020-10-27 [1]
vctrs 0.3.8 2021-04-29 [2]
vegan 2.5-7 2020-11-28 [1]
vipor 0.4.5 2017-03-22 [1]
viridis * 0.6.1 2021-05-11 [1]
viridisLite * 0.4.0 2021-04-13 [1]
withr 2.4.2 2021-04-18 [2]
workflowr * 1.6.2 2020-04-30 [1]
writexl * 1.4.0 2021-04-20 [1]
xfun 0.23 2021-05-15 [1]
XML 3.99-0.6 2021-03-16 [1]
xml2 1.3.2 2020-04-23 [2]
xtable 1.8-4 2019-04-21 [2]
XVector 0.30.0 2020-10-27 [1]
yaml 2.2.1 2020-02-01 [2]
zlibbioc 1.36.0 2020-10-27 [1]
source
CRAN (R 4.0.3)
Bioconductor
Bioconductor
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
Bioconductor
Bioconductor
CRAN (R 4.0.3)
Bioconductor
Bioconductor
Bioconductor
Bioconductor
Bioconductor
Bioconductor
CRAN (R 4.0.2)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.1)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Github (jalvesaq/colorout@79931fd)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
Bioconductor
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
Bioconductor
CRAN (R 4.0.3)
Bioconductor
Bioconductor
Bioconductor
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.1)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.0)
Github (HelenaLC/muscat@c939663)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
Bioconductor
Bioconductor
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.0)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.0)
Bioconductor
Bioconductor
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.1)
Bioconductor
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.1)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.3)
Bioconductor
[1] /pstore/home/macnairw/lib/conda_r3.12
[2] /pstore/home/macnairw/.conda/envs/r_4.0.3/lib/R/library
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-conda-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /pstore/home/macnairw/.conda/envs/r_4.0.3/lib/libopenblasp-r0.3.12.so
locale:
[1] LC_CTYPE=C LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] grid parallel stats4 stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] rmarkdown_2.8 writexl_1.4.0
[3] readxl_1.3.1 ComplexHeatmap_2.6.2
[5] fgsea_1.16.0 tictoc_1.0.1
[7] reshape2_1.4.4 scater_1.18.6
[9] Matrix.utils_0.9.8 seriation_1.2-9
[11] UpSetR_1.4.0 dplyr_1.0.6
[13] readr_1.4.0 tidyr_1.1.3
[15] tibble_3.1.2 tidyverse_1.3.1
[17] rtracklayer_1.50.0 ggrepel_0.9.1
[19] reticulate_1.20 MASS_7.3-54
[21] phyloseq_1.34.0 ANCOMBC_1.0.5
[23] purrr_0.3.4 patchwork_1.1.1
[25] nnls_1.4 muscat_1.5.1
[27] DropletUtils_1.10.3 edgeR_3.32.1
[29] limma_3.46.0 BiocParallel_1.24.1
[31] Matrix_1.3-3 scran_1.18.7
[33] SingleCellExperiment_1.12.0 SummarizedExperiment_1.20.0
[35] Biobase_2.50.0 GenomicRanges_1.42.0
[37] GenomeInfoDb_1.26.7 IRanges_2.24.1
[39] S4Vectors_0.28.1 BiocGenerics_0.36.1
[41] MatrixGenerics_1.2.1 matrixStats_0.59.0
[43] googlesheets_0.3.0 forcats_0.5.1
[45] ggplot2_3.3.3 scales_1.1.1
[47] viridis_0.6.1 viridisLite_0.4.0
[49] assertthat_0.2.1 stringr_1.4.0
[51] data.table_1.14.0 magrittr_2.0.1
[53] circlize_0.4.12 RColorBrewer_1.1-2
[55] BiocStyle_2.18.1 colorout_1.2-2
[57] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] R.methodsS3_1.8.1 bit64_4.0.5
[3] knitr_1.33 irlba_2.3.3
[5] DelayedArray_0.16.3 R.utils_2.10.1
[7] RCurl_1.98-1.3 doParallel_1.0.16
[9] generics_0.1.0 callr_3.7.0
[11] microbiome_1.12.0 usethis_2.0.1
[13] RSQLite_2.2.7 future_1.21.0
[15] bit_4.0.4 xml2_1.3.2
[17] lubridate_1.7.10 httpuv_1.6.1
[19] xfun_0.23 hms_1.1.0
[21] jquerylib_0.1.4 TSP_1.1-10
[23] evaluate_0.14 promises_1.2.0.1
[25] fansi_0.4.2 progress_1.2.2
[27] caTools_1.18.2 dbplyr_2.1.1
[29] igraph_1.2.6 DBI_1.1.1
[31] geneplotter_1.68.0 ellipsis_0.3.2
[33] backports_1.2.1 permute_0.9-5
[35] annotate_1.68.0 sparseMatrixStats_1.2.1
[37] vctrs_0.3.8 remotes_2.3.0
[39] Cairo_1.5-12.2 cachem_1.0.5
[41] withr_2.4.2 grr_0.9.5
[43] sctransform_0.3.2 vegan_2.5-7
[45] GenomicAlignments_1.26.0 prettyunits_1.1.1
[47] cluster_2.1.2 ape_5.5
[49] crayon_1.4.1 genefilter_1.72.1
[51] labeling_0.4.2 pkgconfig_2.0.3
[53] pkgload_1.2.1 nlme_3.1-152
[55] vipor_0.4.5 devtools_2.4.1
[57] blme_1.0-5 rlang_0.4.11
[59] globals_0.14.0 lifecycle_1.0.0
[61] registry_0.5-1 modelr_0.1.8
[63] rsvd_1.0.5 cellranger_1.1.0
[65] rprojroot_2.0.2 Rhdf5lib_1.12.1
[67] boot_1.3-28 reprex_2.0.0
[69] beeswarm_0.3.1 processx_3.5.2
[71] GlobalOptions_0.1.2 png_0.1-7
[73] rjson_0.2.20 bitops_1.0-7
[75] R.oo_1.24.0 KernSmooth_2.23-20
[77] rhdf5filters_1.2.1 Biostrings_2.58.0
[79] blob_1.2.1 DelayedMatrixStats_1.12.3
[81] shape_1.4.6 parallelly_1.25.0
[83] beachmat_2.6.4 memoise_2.0.0
[85] plyr_1.8.6 gplots_3.1.1
[87] zlibbioc_1.36.0 compiler_4.0.3
[89] dqrng_0.3.0 clue_0.3-59
[91] lme4_1.1-27 DESeq2_1.30.1
[93] snakecase_0.11.0 cli_2.5.0
[95] Rsamtools_2.6.0 ade4_1.7-16
[97] XVector_0.30.0 lmerTest_3.1-3
[99] listenv_0.8.0 ps_1.6.0
[101] TMB_1.7.20 mgcv_1.8-35
[103] tidyselect_1.1.1 stringi_1.6.2
[105] highr_0.9 yaml_2.2.1
[107] BiocSingular_1.6.0 locfit_1.5-9.4
[109] sass_0.4.0 fastmatch_1.1-0
[111] tools_4.0.3 future.apply_1.7.0
[113] rstudioapi_0.13 bluster_1.0.0
[115] foreach_1.5.1 git2r_0.28.0
[117] janitor_2.1.0 gridExtra_2.3
[119] farver_2.1.0 Rtsne_0.15
[121] digest_0.6.27 BiocManager_1.30.15
[123] Rcpp_1.0.6 broom_0.7.6
[125] scuttle_1.0.4 later_1.2.0
[127] httr_1.4.2 AnnotationDbi_1.52.0
[129] Rdpack_2.1.1 colorspace_2.0-1
[131] rvest_1.0.0 XML_3.99-0.6
[133] fs_1.5.0 splines_4.0.3
[135] statmod_1.4.36 multtest_2.46.0
[137] sessioninfo_1.1.1 xtable_1.8-4
[139] jsonlite_1.7.2 nloptr_1.2.2.2
[141] testthat_3.0.2 R6_2.5.0
[143] pillar_1.6.1 htmltools_0.5.1.1
[145] glue_1.4.2 fastmap_1.1.0
[147] minqa_1.2.4 BiocNeighbors_1.8.2
[149] codetools_0.2-18 pkgbuild_1.2.0
[151] utf8_1.2.1 lattice_0.20-44
[153] bslib_0.2.5 numDeriv_2016.8-1.1
[155] pbkrtest_0.5.1 ggbeeswarm_0.6.0
[157] colorRamps_2.3 gtools_3.8.2
[159] survival_3.2-11 glmmTMB_1.0.2.1
[161] desc_1.3.0 biomformat_1.18.0
[163] munsell_0.5.0 GetoptLong_1.0.5
[165] rhdf5_2.34.0 GenomeInfoDbData_1.2.4
[167] iterators_1.0.13 HDF5Array_1.18.1
[169] variancePartition_1.20.0 haven_2.4.1
[171] gtable_0.3.0 rbibutils_2.1.1