MS: Double-checking selected markers
Will Macnair
Institute for Molecular Life Sciences, University of Zurich, SwitzerlandSwiss Institute of Bioinformatics (SIB), University of Zurich, SwitzerlandAugust 26, 2021
Last updated: 2021-08-26
Checks: 4 3
Knit directory: MS_lesions/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
The global environment had objects present when the code in the R Markdown file was run. These objects can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment. Use wflow_publish or wflow_build to ensure that the code is always run in an empty environment.
The following objects were defined in the global environment when these results were created:
| Name | Class | Size |
|---|---|---|
| q | function | 1008 bytes |
The command set.seed(20210118) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
- calc_conos_on_panel
- calc_expression_cartana
- calc_expression_roche
- calc_median_counts_cartana
- calc_median_counts_roche
- calc_muscat_on_clusters
- calc_n_expressed
- calc_random_forest
- load_agreed_markers
- load_conos
- load_micro_macro_markers
- load_pseudobulk
- load_qc
- load_umap_many_dt
- plot_astro_dotplot
- plot_classifier_heatmaps
- plot_dotplot_broad
- plot_dotplot_dheeraj
- plot_dotplot_dheeraj_compact
- plot_dotplot_fine
- plot_dotplot_micro_ma
- plot_gsea_results_all
- plot_gsea_results_ctrl
- plot_marker_heatmaps_cartana
- plot_marker_heatmaps_roche
- plot_muscat_dotplot
- plot_muscat_dotplot_all
- plot_muscat_dotplot_ctrl
- plot_muscat_results_all
- plot_muscat_results_ctrl
- plot_n_expressed_distributions
- plot_oligo_dotplot
- plot_panel_conos_clusters
- plot_panel_conos_umap_densities
- plot_panel_conos_umaps
- plot_pb_qc
- plot_umap_final_celltypes
- plot_umap_final_celltypes_sel
- plot_umap_final_celltypes_sel_broad
- save_outputs
- session_info
- session-info-chunk-inserted-by-workflowr
- setup_input
- setup_outputs
To ensure reproducibility of the results, delete the cache directory ms12_markers_cache and re-run the analysis. To have workflowr automatically delete the cache directory prior to building the file, set delete_cache = TRUE when running wflow_build() or wflow_publish().
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 96d385e. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rprofile
Ignored: .Rproj.user/
Ignored: ._.DS_Store
Ignored: ._MS_lesions.sublime-project
Ignored: ._heatmap_check.png
Ignored: ._metadata_checked_20210712 - subject_metadata.csv
Ignored: ._model_example_fix.png
Ignored: .log/
Ignored: MS_lesions.sublime-project
Ignored: MS_lesions.sublime-workspace
Ignored: analysis/.__site.yml
Ignored: analysis/fig_muscat_cache/
Ignored: analysis/ms02_doublet_id_cache/
Ignored: analysis/ms03_SampleQC_cache/
Ignored: analysis/ms04_conos_cache/
Ignored: analysis/ms05_splitting_cache/
Ignored: analysis/ms06_sccaf_cache/
Ignored: analysis/ms07_soup_cache/
Ignored: analysis/ms08_modules_cache/
Ignored: analysis/ms09_ancombc_cache/
Ignored: analysis/ms09_ancombc_clean_1e3_cache/
Ignored: analysis/ms09_ancombc_clean_2e3_cache/
Ignored: analysis/ms10_muscat_run01_cache/
Ignored: analysis/ms10_muscat_run02_cache/
Ignored: analysis/ms10_muscat_template_broad_cache/
Ignored: analysis/ms10_muscat_template_fine_cache/
Ignored: analysis/ms11_paga_cache/
Ignored: analysis/ms12_markers_cache/
Ignored: analysis/ms13_labelling_cache/
Ignored: analysis/ms14_lesions_cache/
Ignored: analysis/ms15_mofa_sample_gm_cache/
Ignored: analysis/ms15_mofa_sample_gm_superclean_cache/
Ignored: analysis/ms15_mofa_sample_wm_cache/
Ignored: analysis/ms15_mofa_sample_wm_superclean_cache/
Ignored: analysis/ms15_patients_cache/
Ignored: analysis/ms15_patients_gm_cache/
Ignored: analysis/ms15_patients_sample_level_cache/
Ignored: analysis/ms15_patients_w_ms_cache/
Ignored: analysis/supp06_sccaf_cache/
Ignored: analysis/supp07_superclean_check_cache/
Ignored: analysis/supp09_ancombc_cache/
Ignored: analysis/supp10_muscat_cache/
Ignored: analysis/supp10_muscat_ctrl_gm_vs_wm_cache/
Ignored: analysis/supp10_muscat_heatmaps_cache/
Ignored: analysis/supp10_muscat_olg_pc1_cache/
Ignored: analysis/supp10_muscat_olg_pc2_cache/
Ignored: analysis/supp10_muscat_olg_pc_cache/
Ignored: analysis/supp10_muscat_regression_cache/
Ignored: analysis/supp10_muscat_soup_cache/
Ignored: code/.recovery/
Ignored: code/muscat_plan.txt
Ignored: data/
Ignored: figures/
Ignored: output/
Ignored: tmp/
Untracked files:
Untracked: code/dev_check_pmi_distns_20210824.R
Untracked: code/dev_redo_gsea_20210806.R
Untracked: code/dev_sketch_pseudobulk_2021-08-19.R
Unstaged changes:
Modified: analysis/ms08_modules.Rmd
Modified: analysis/ms09_ancombc.Rmd
Modified: analysis/ms12_markers.Rmd
Modified: analysis/ms14_lesions.Rmd
Modified: analysis/supp10_muscat_regression.Rmd
Modified: code/dev_edger_on_mofa_20210804.R
Modified: code/ms08_modules.R
Modified: code/ms09_ancombc.R
Modified: code/ms12_markers.R
Modified: code/ms14_lesions.R
Modified: code/ms15_mofa.R
Modified: code/supp10_muscat.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ms12_markers.Rmd) and HTML (docs/ms12_markers.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 6f577cc | Macnair | 2021-08-24 | Tweak dotplot in ms12_markers |
| html | 6f577cc | Macnair | 2021-08-24 | Tweak dotplot in ms12_markers |
| Rmd | f60e5c1 | Macnair | 2021-08-03 | Add astrocyte layer genes to ms12_markers |
| html | f60e5c1 | Macnair | 2021-08-03 | Add astrocyte layer genes to ms12_markers |
| Rmd | c20e65d | Macnair | 2021-07-02 | Add UMAP annotated with fine celltypes to ms12_markers |
| html | 0ec42e1 | Macnair | 2021-06-20 | Update GSEA analysis, heatmaps, lesion-specific |
| html | be76f21 | Macnair | 2021-06-10 | Add Dheeraj’s genes to dotplots |
| Rmd | 0ee7fc4 | Macnair | 2021-06-08 | Add Dheeraj’s genes to ms12_markers |
| html | 0ee7fc4 | Macnair | 2021-06-08 | Add Dheeraj’s genes to ms12_markers |
| Rmd | 1ea21aa | Macnair | 2021-06-03 | Updated marker analysis |
| html | 1ea21aa | Macnair | 2021-06-03 | Updated marker analysis |
| Rmd | 768da0e | Macnair | 2021-05-27 | Marker panel validation |
| html | 768da0e | Macnair | 2021-05-27 | Marker panel validation |
| Rmd | 58205c2 | Macnair | 2021-05-21 | Update with random effects and markers |
| html | 58205c2 | Macnair | 2021-05-21 | Update with random effects and markers |
| html | 9852840 | Macnair | 2021-05-12 | Adding docs to repo for first time - massive! |
Setup / definitions
Libraries
Helper functions
source('code/ms00_utils.R')
source('code/ms03_SampleQC.R')
source('code/ms04_conos.R')
source('code/ms07_soup.R')
source('code/ms10_muscat_runs.R')
source('code/ms12_markers.R')Inputs
# base inputs
sce_f = 'data/sce_raw/ms_sce.rds'
labels_f = 'data/byhand_markers/validation_markers_2021-05-31.csv'
labelled_f = 'output/ms13_labelling/conos_labelled_2021-05-31.txt.gz'
meta_f = 'data/metadata/metadata_updated_20201127.txt'
# qc info
qc_dir = 'output/ms03_SampleQC'
qc_f = file.path(qc_dir, 'ms_qc_dt.txt')
# define hand-selected marker location
byhand_f = paste0("data/byhand_markers/",
"Copy of Copy of Marker_selection_for_validation_MS_snucseq_30102020",
" Ediinburgh.xlsx - final markers for Cartana panel.csv")
roche_f = paste0("data/byhand_markers/",
"Copy of Copy of Marker_selection_for_validation_MS_snucseq_30102020",
" Ediinburgh.xlsx - analysis_Roche_April2021.csv")
# umap embedding options
umap_many_f = 'output/ms04_conos/conos_umap_sub_2021-02-11.txt'
# define pseudobulk files
soup_dir = 'output/ms07_soup'
date_soup = '2021-06-01'
pb_fine_f = sprintf('%s/pb_sum_fine_%s.rds', soup_dir, date_soup)
prop_fine_f = sprintf('%s/pb_prop_fine_%s.rds', soup_dir, date_soup)
pb_broad_f = file.path(soup_dir, 'pb_sum_broad_2021-06-01.rds')
pb_soup_f = file.path(soup_dir, 'pb_soup_broad_maximum_2021-06-01.rds')Outputs
# set up directory
save_dir = 'output/ms12_markers'
date_tag = '2021-06-02'
if (!dir.exists(save_dir))
dir.create(save_dir)
medians_pat = sprintf('%s/median_counts_%s_%s.txt', save_dir, '%s', date_tag)
prop_exp_pat = sprintf('%s/prop_exp_%s_%s.txt', save_dir, '%s', date_tag)
n_exp_f = sprintf('%s/n_exp_%s.txt', save_dir, date_tag)
conf_dt_f = sprintf('%s/confusion_matrix_dt_%s.txt', save_dir, date_tag)
conos_chk_f = sprintf('%s/conos_chk_%s.txt', save_dir, date_tag)
# group some celltypes together
broad_list = list(
`OPCs / COPs` = 'OPCs / COPs',
Oligodendrocytes = 'Oligodendrocytes',
`Astrocytes`='Astrocytes',
`Microglia + Immune`=c('Microglia', 'Immune'),
`Excitatory neurons`='Excitatory neurons',
`Inhibitory neurons`='Inhibitory neurons',
`Endothelial + Pericytes`=c('Endothelial cells', 'Pericytes')
)
min_cells = 10
fgsea_cut = 0.1
muscat_ctrl_f = sprintf('%s/muscat_ctrl_markers_%s.txt', save_dir, '2021-07-27')
fgsea_ctrl_f = sprintf('%s/muscat_ctrl_gsea_%s.txt', save_dir, '2021-07-27')
muscat_all_f = sprintf('%s/muscat_markers_%s.txt', save_dir, '2021-07-27')
fgsea_all_f = sprintf('%s/muscat_gsea_%s.txt', save_dir, '2021-07-27')Load inputs
# do pseudobulking
pb_fine = pb_fine_f %>% readRDS
props_fine = prop_fine_f %>% readRDS
# load soup stuff
pb_soup = pb_soup_f %>% readRDS
pb_broad = pb_broad_f %>% readRDS
contam_dt = .calc_contam_dt(pb_soup, pb_broad, min_cells)
rm(pb_soup, pb_broad)
biotypes_dt = .get_biotypes_dt(gtf_f)
labels_dt = load_names_dt(labels_f) %>%
.[, conos := NULL]cols_dt = colData(pb_fine) %>% as.data.frame %>%
as.data.table(keep.rownames = 'sample_id') %>%
.[, .(sample_id, patient_id, lesion_type)]
conos_dt = load_labelled_dt(labelled_f, labels_f) %>%
merge(cols_dt, by = 'sample_id')pb_qc_dt = calc_pb_qc_dt(qc_dir, qc_f)# get genes from panel
byhand_ls = load_byhand_genes(byhand_f, labels_dt, pb_fine)
byhand_gs = map(byhand_ls, ~.x$symbol) %>% unlist %>% unique
# get genes from roche panel
roche_dt = load_roche_genes(roche_f, labels_dt, pb_fine)
roche_gs = roche_dt$symbol %>% unique
# get genes from Dheeraj's list
dheeraj_dt = load_dheeraj_dt(pb_fine)micro_gs = load_micro_macro_markers()final_cols = make_final_cols(labels_dt)
umap_dt = load_umap_many(umap_many_f, conos_dt)Processing / calculations
n_exp_dt = calc_n_exp_dt(n_exp_f, sce_f, conos_dt, byhand_ls)prop_exp_f = sprintf(prop_exp_pat, 'cartana')
prop_exp_dt = calc_prop_exp_dt(prop_exp_f, sce_f, conos_dt, byhand_ls)
seriate_obj = calc_heatmap_ordering(prop_exp_dt)prop_exp_f = sprintf(prop_exp_pat, 'roche')
prop_exp_dt_roche = calc_prop_exp_dt(prop_exp_f, sce_f, conos_dt, list(roche_dt))
seriate_roche = calc_heatmap_ordering(prop_exp_dt_roche)medians_f = sprintf(medians_pat, 'cartana')
medians_dt = calc_medians_dt(medians_f, sce_f, conos_dt, byhand_ls)medians_f = sprintf(medians_pat, 'roche')
medians_dt_roche = calc_medians_dt(medians_f, sce_f, conos_dt, list(roche_dt))set.seed(123)
conf_dt = test_random_forest(conf_dt_f, sce_f, conos_dt,
min_exp_per_cell = 10, n_per_conos = 1000, n_cores = 16)set.seed(123)
conos_chk = calc_conos_on_panel(sce_f, conos_dt, byhand_gs, conos_chk_f)# get muscat markers within healthy samples
muscat_ctrl = calc_muscat_on_clusters(pb_fine, labels_dt,
contam_dt, biotypes_dt, broad_list, muscat_ctrl_f, sel_lesions = c('WM', 'GM'),
min_cells = 10, n_cores = 8)
fgsea_ctrl = calc_fgsea_on_muscat(muscat_ctrl, labels_dt, fgsea_ctrl_f,
fgsea_cut = 0.1, n_cores = 8)
# get muscat markers within all samples
muscat_all = calc_muscat_on_clusters(pb_fine, labels_dt,
contam_dt, biotypes_dt, broad_list, muscat_all_f, sel_lesions = c('WM', 'GM'),
min_cells = 10, n_cores = 8)
fgsea_all = calc_fgsea_on_muscat(muscat_all, labels_dt, fgsea_all_f,
fgsea_cut = 0.1, n_cores = 8)Analysis
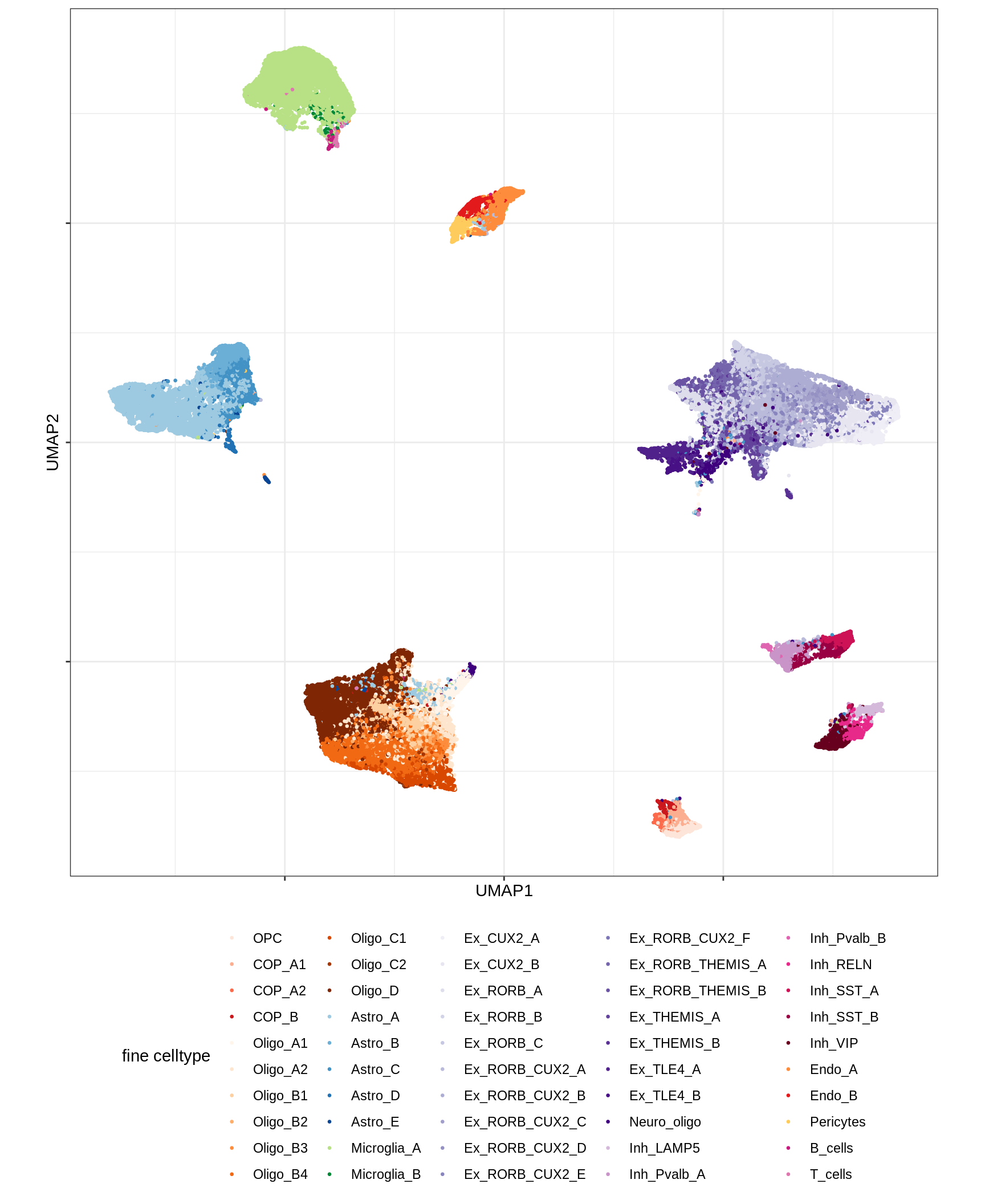
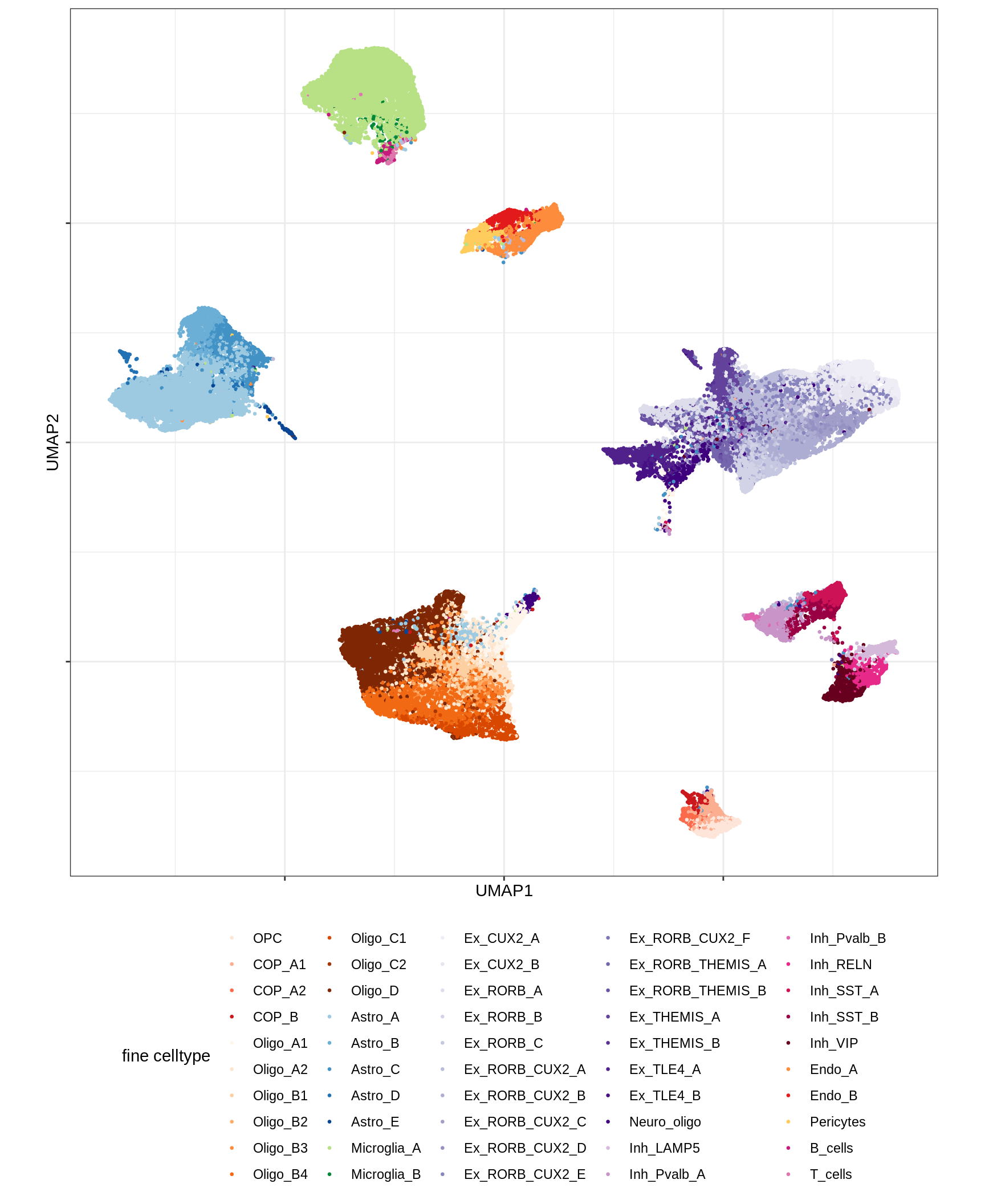
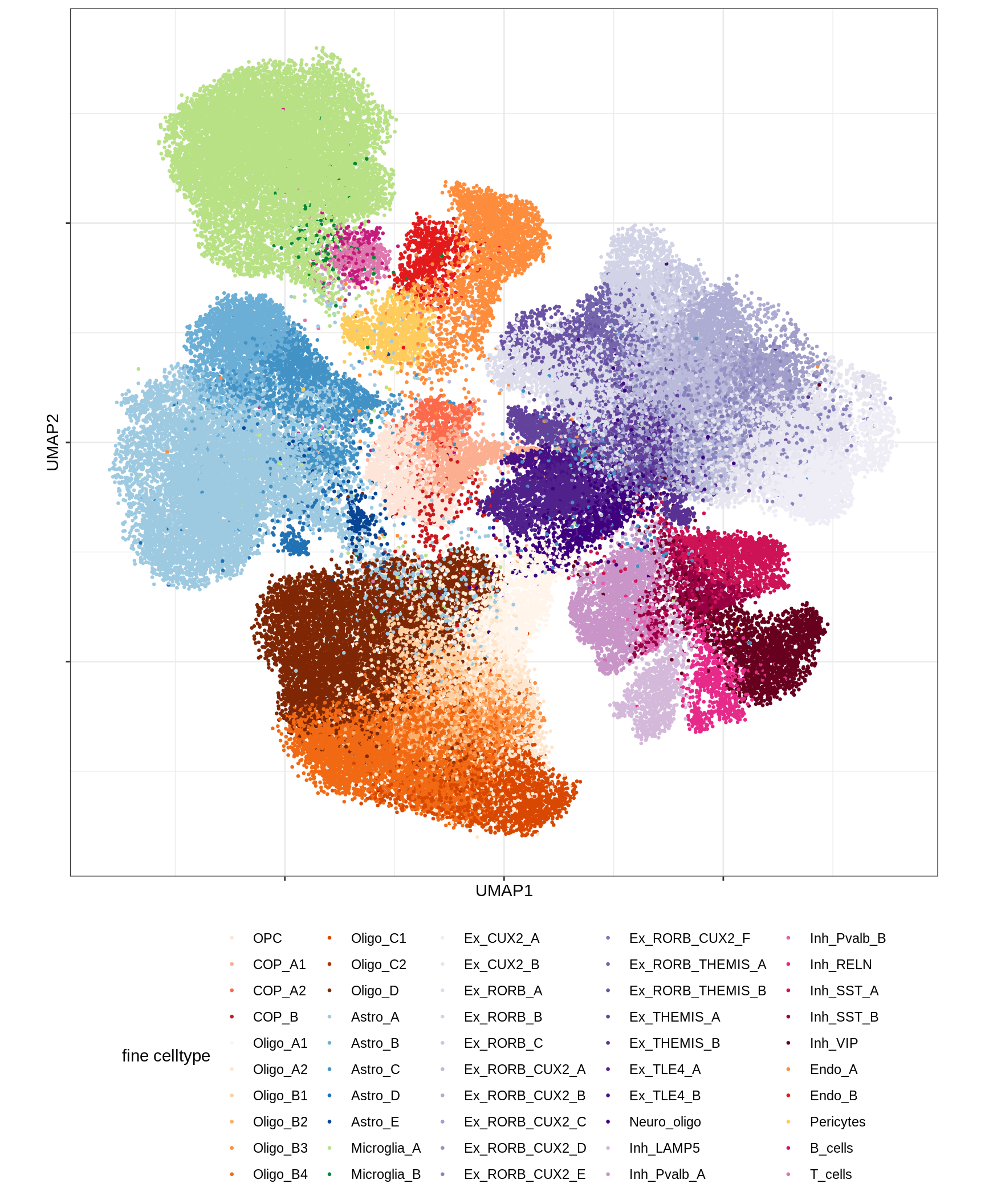
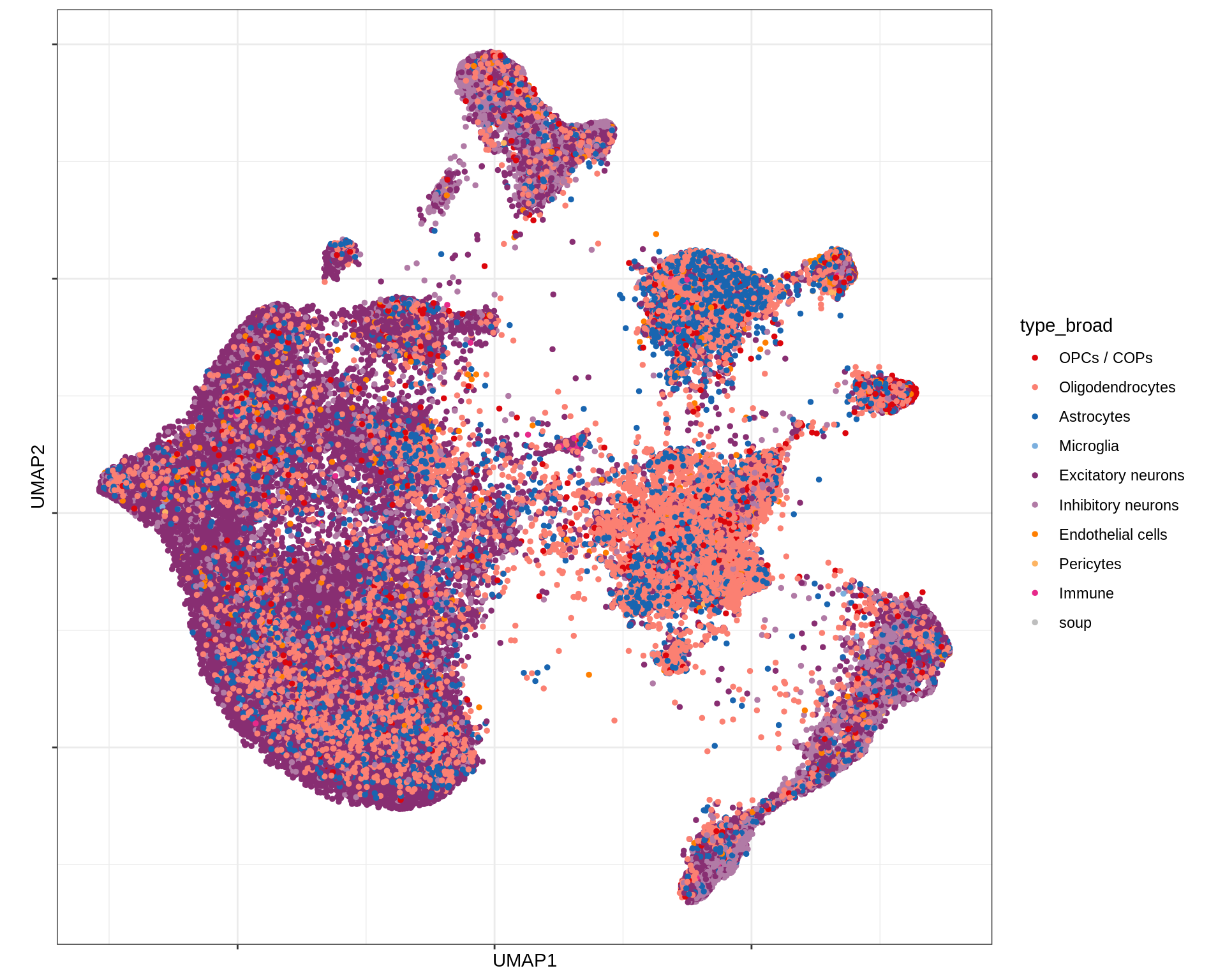
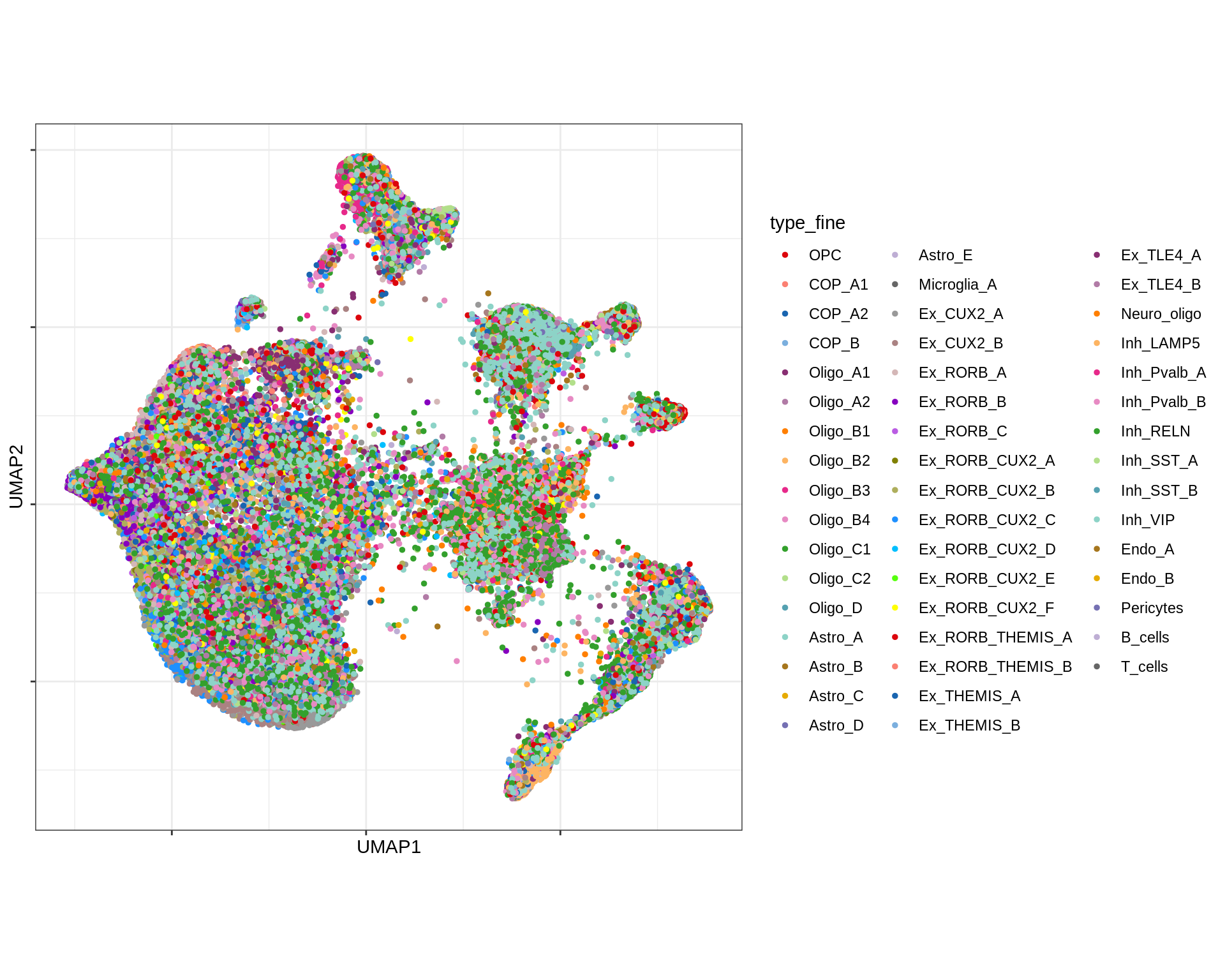
UMAP of final celltypes
message('min_dist = 1, spread = 2')min_dist = 1, spread = 2(plot_umap_final_celltypes(umap_dt[ min_dist == 1 & spread == 2 ],
final_cols))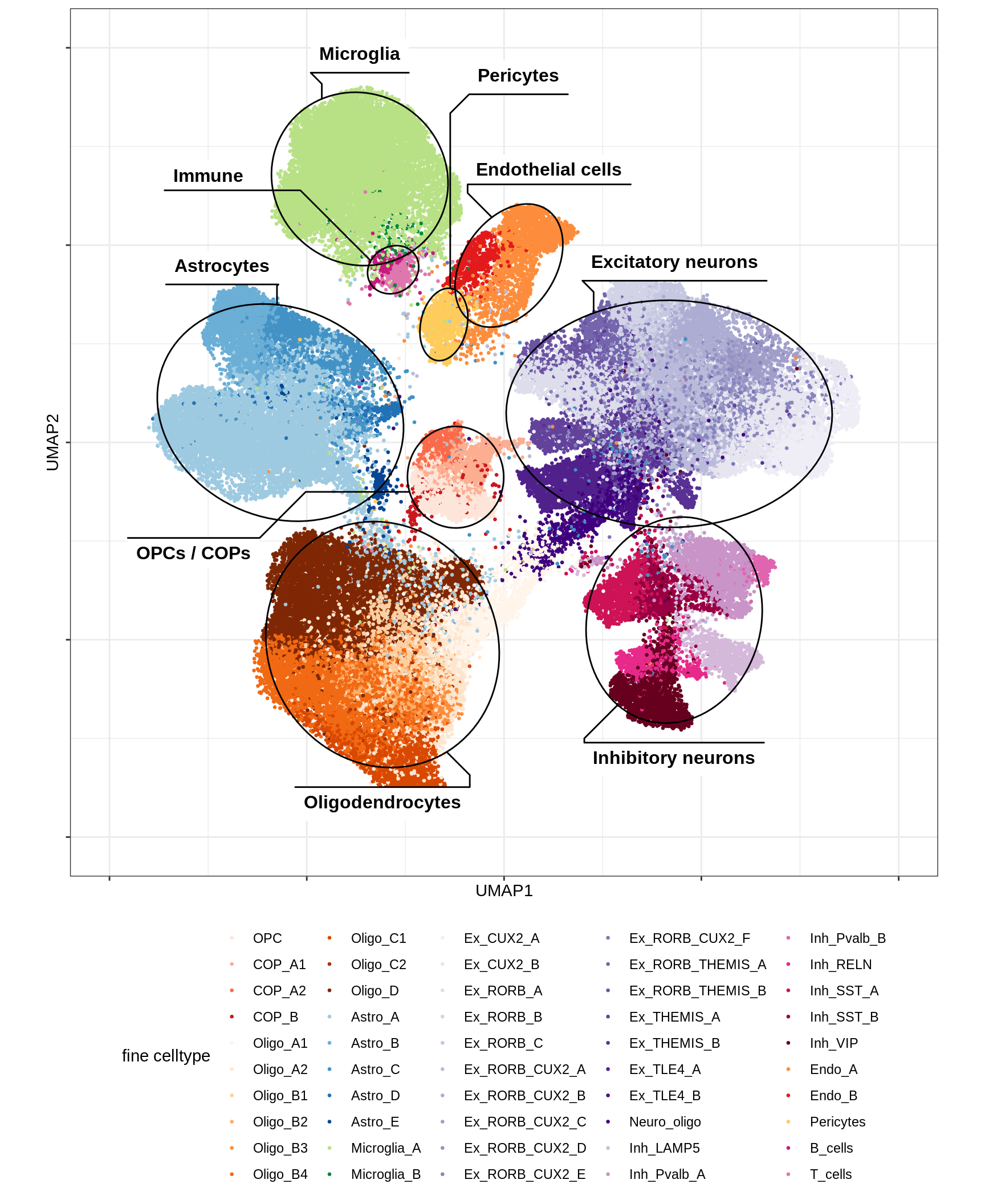
| Version | Author | Date |
|---|---|---|
| 0ec42e1 | Macnair | 2021-06-20 |
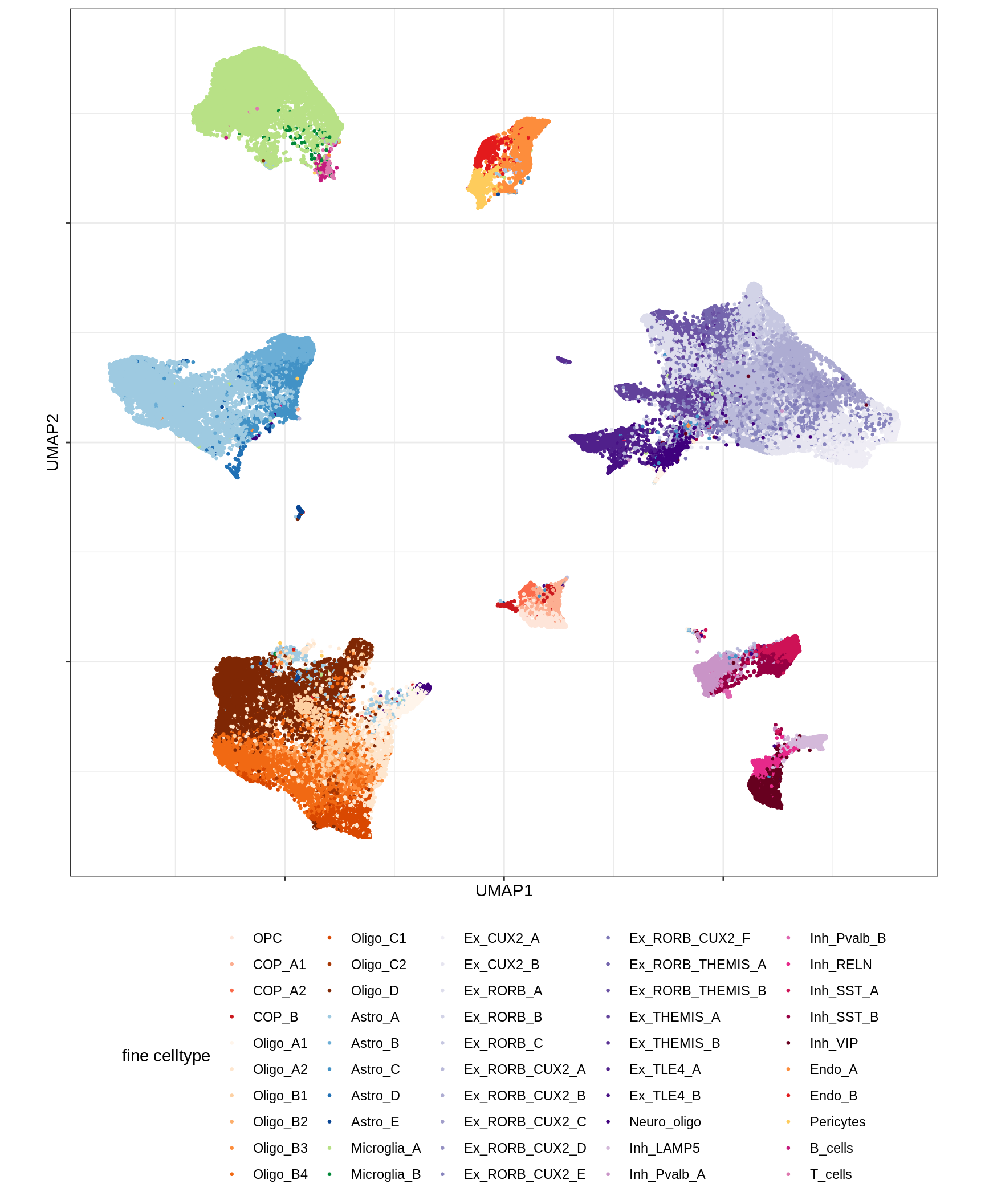
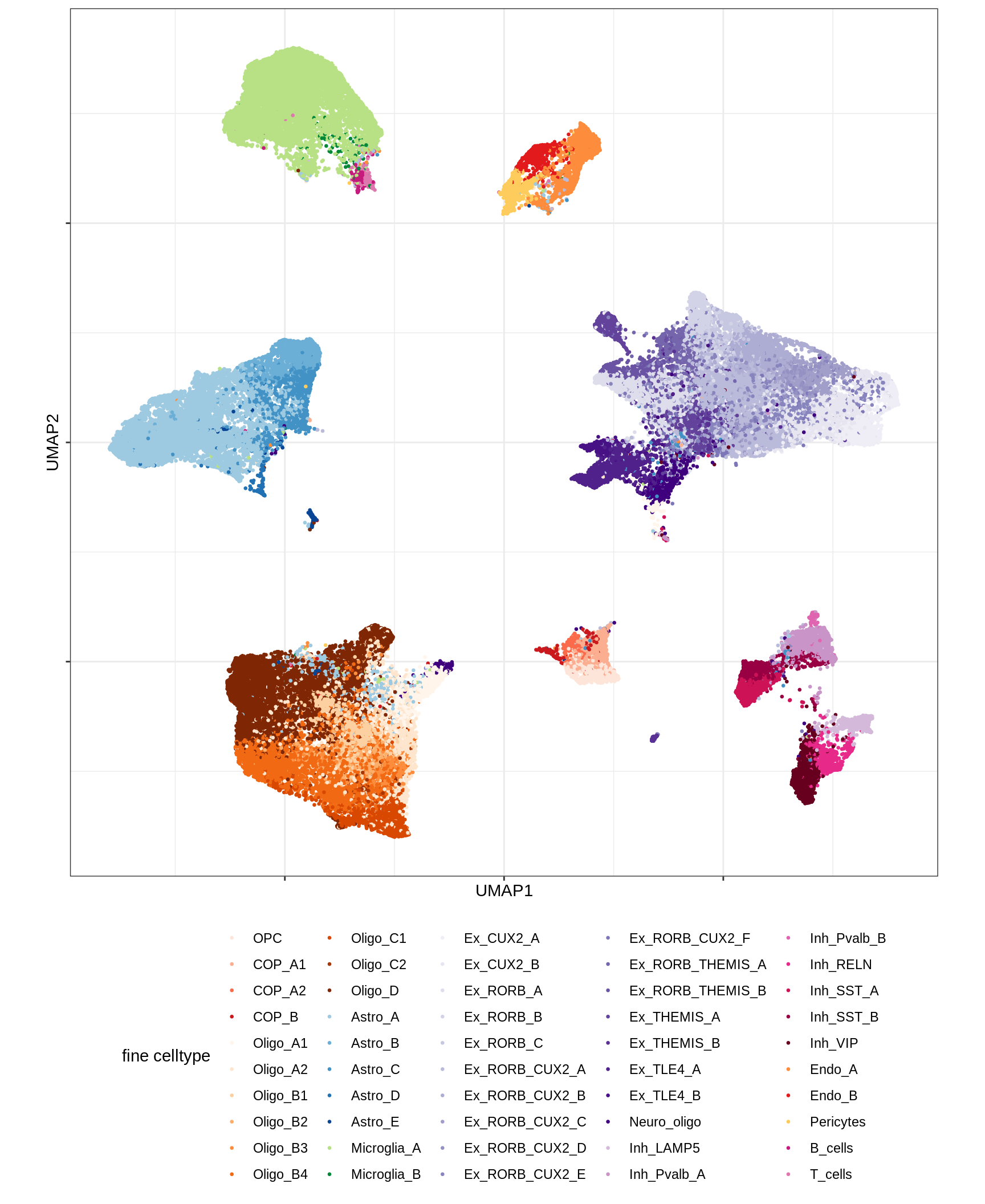
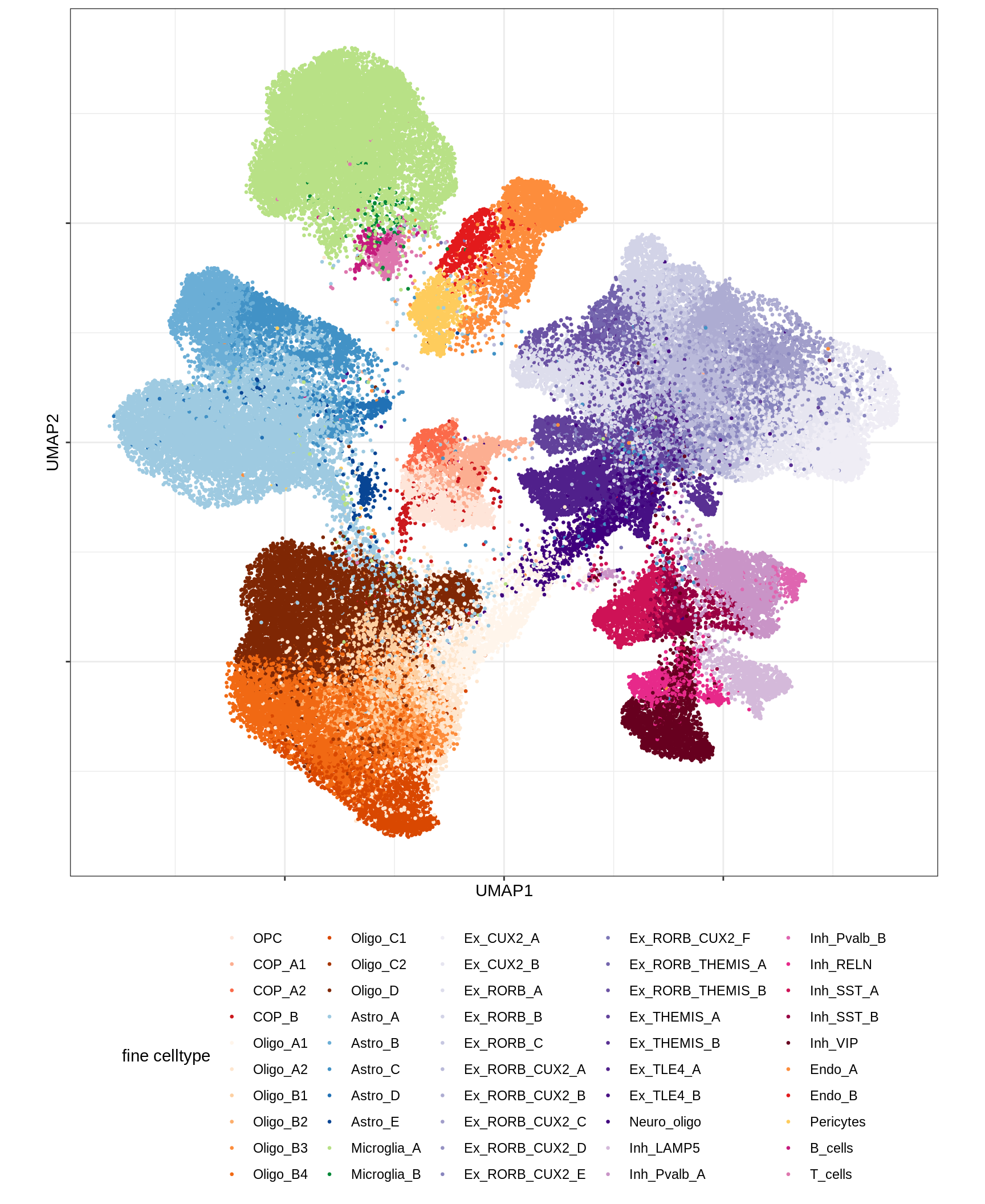
UMAP of final celltypes - broad
message('min_dist = 1, spread = 2')min_dist = 1, spread = 2(plot_umap_final_celltypes_broad(umap_dt[ min_dist == 1 & spread == 2 ]))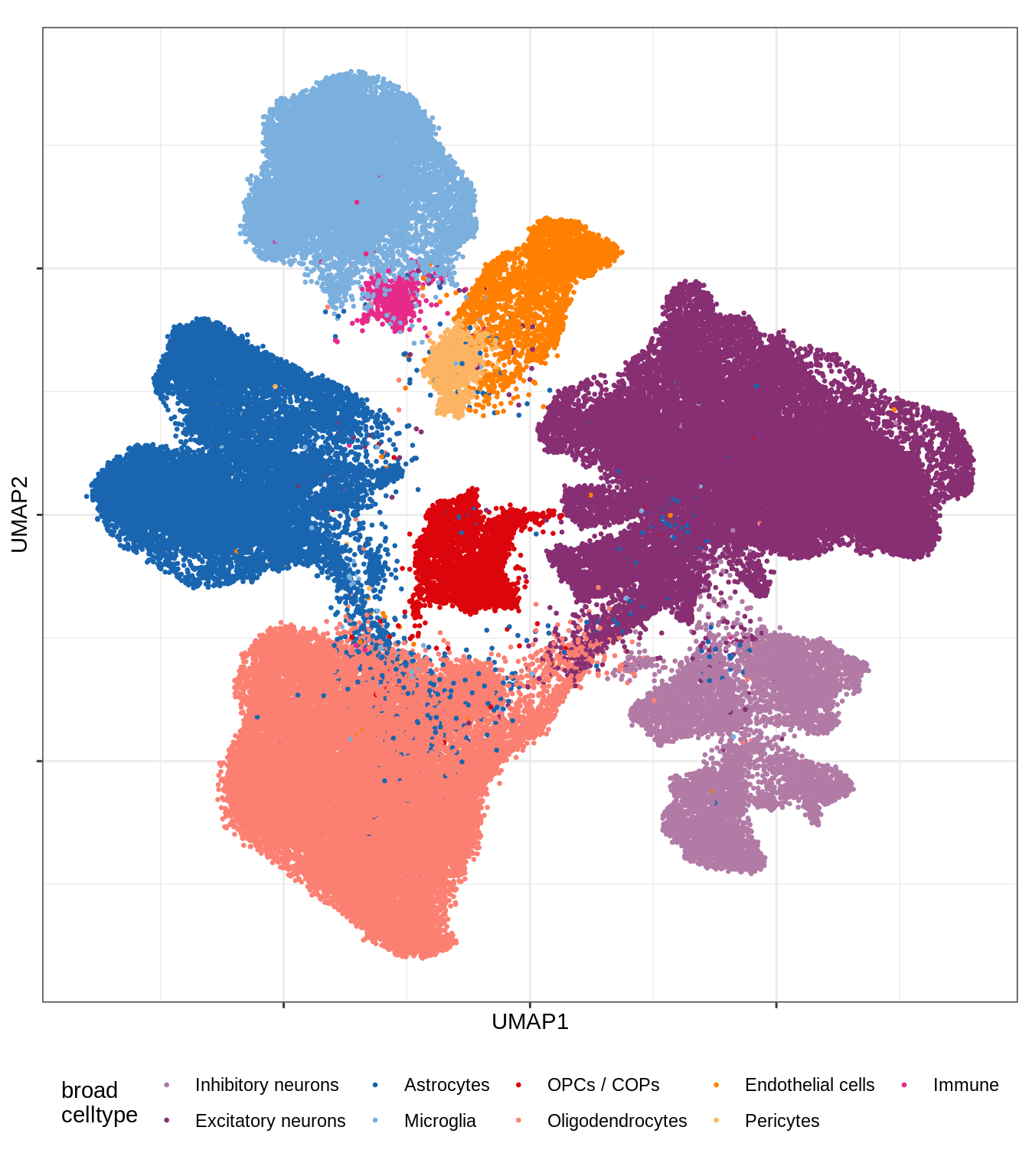
UMAP of OPCs + oligos only
message('min_dist = 1, spread = 2')min_dist = 1, spread = 2umap_sub = umap_dt[ min_dist == 1 & spread == 2 ] %>%
.[ type_broad %in% c('Oligodendrocytes', 'OPCs / COPs') ]
(plot_umap_fine(umap_sub, final_cols, by_var = 'type_fine',
xlim = c(0.1, 0.6), ylim = c(0, 0.5)))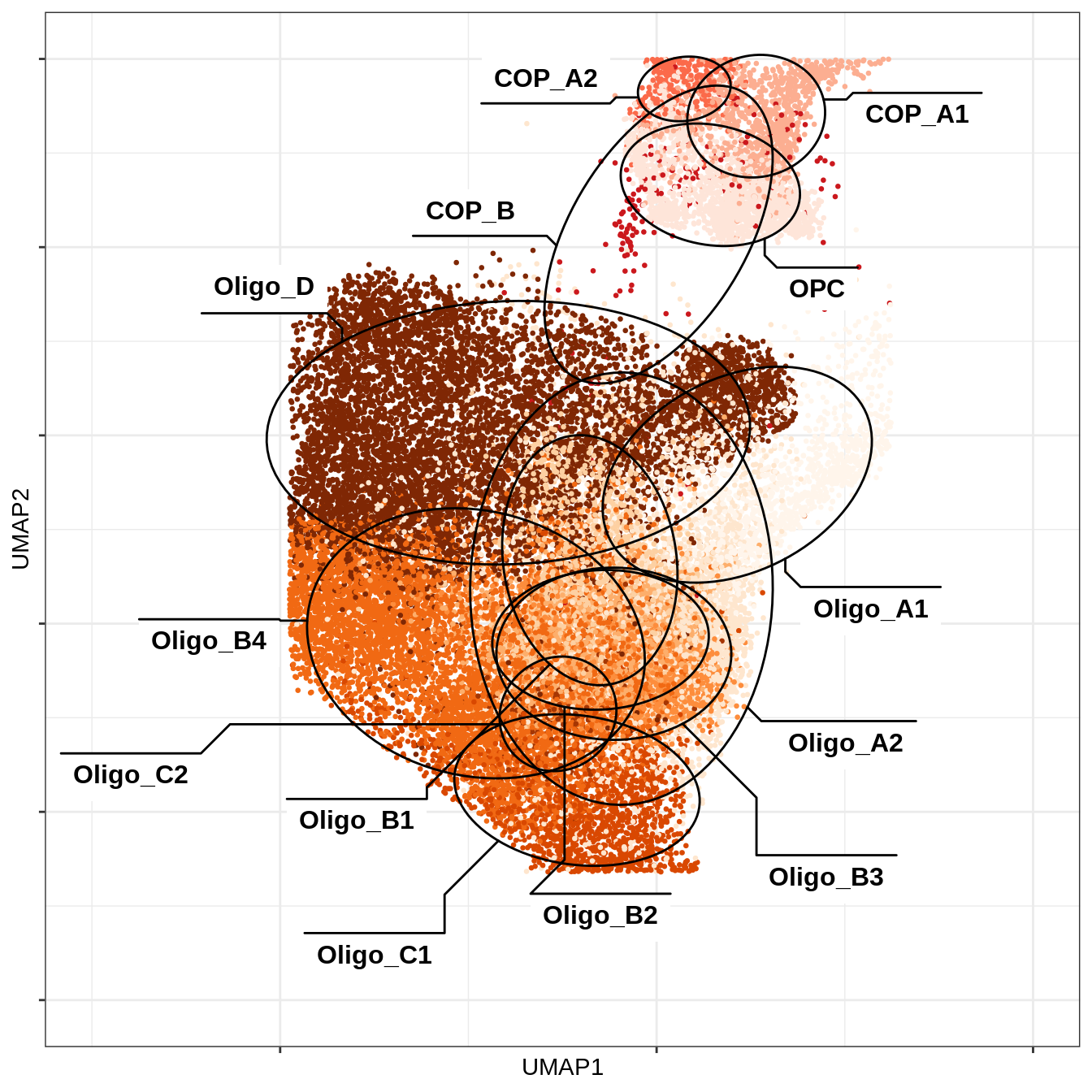
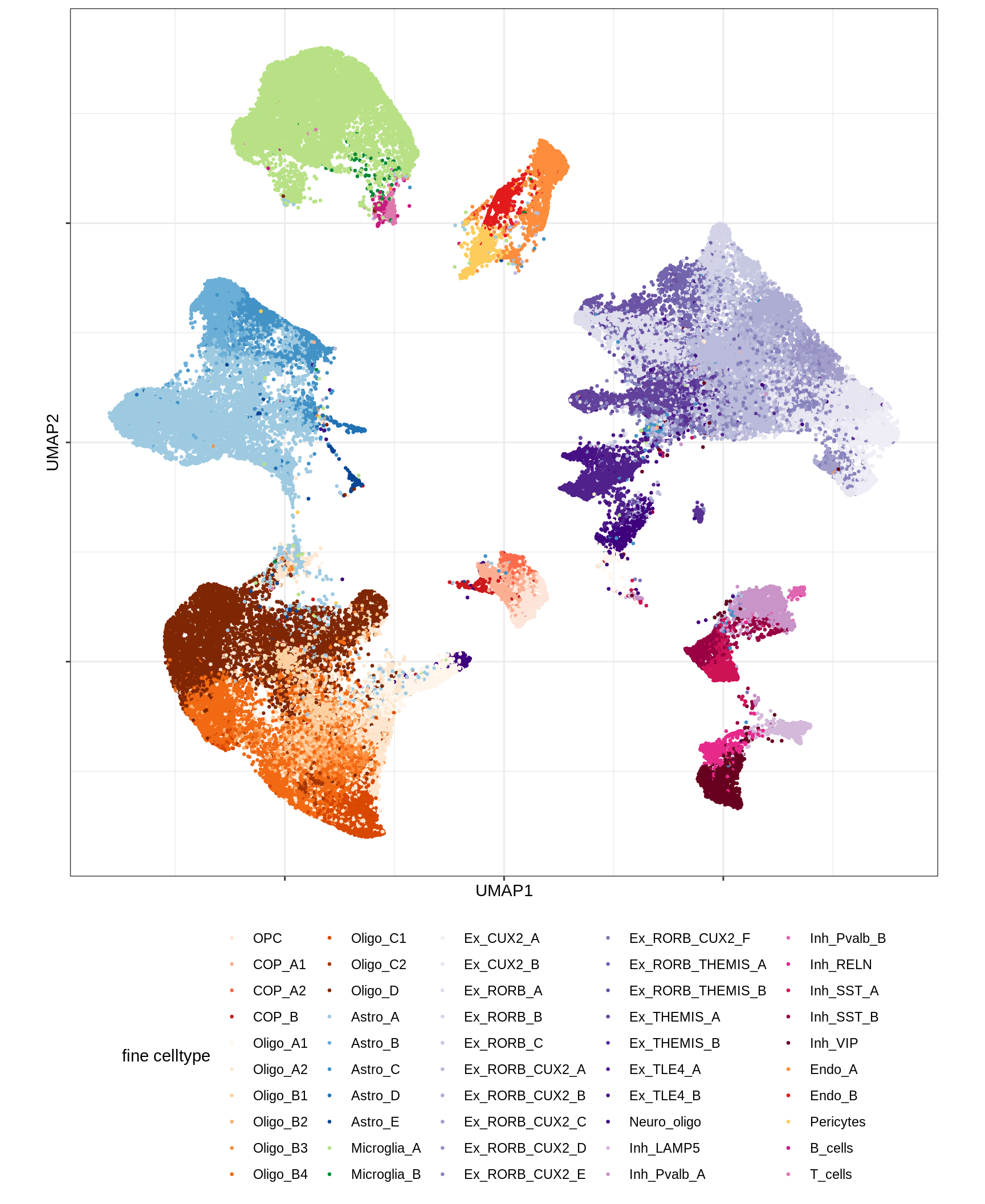
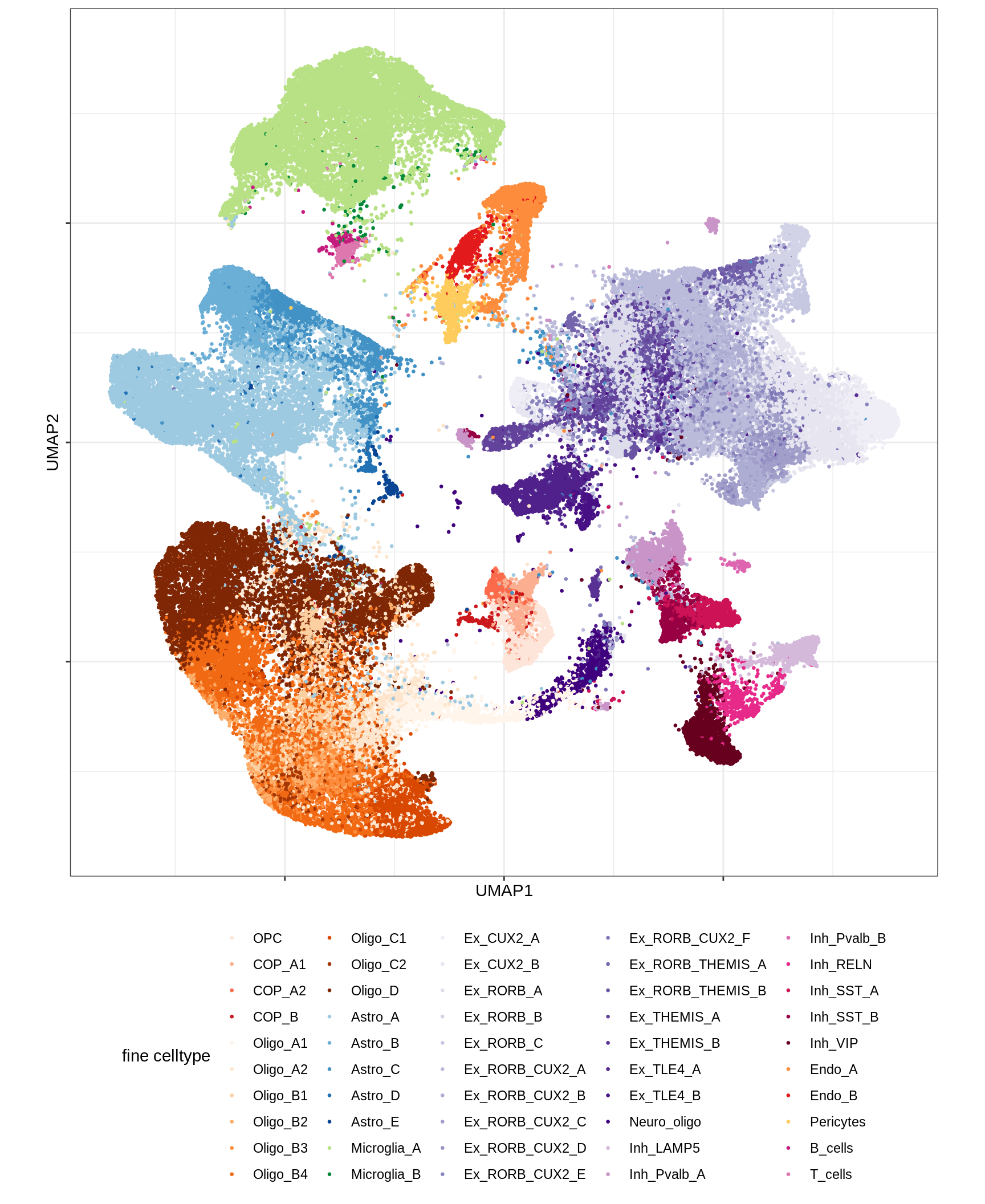
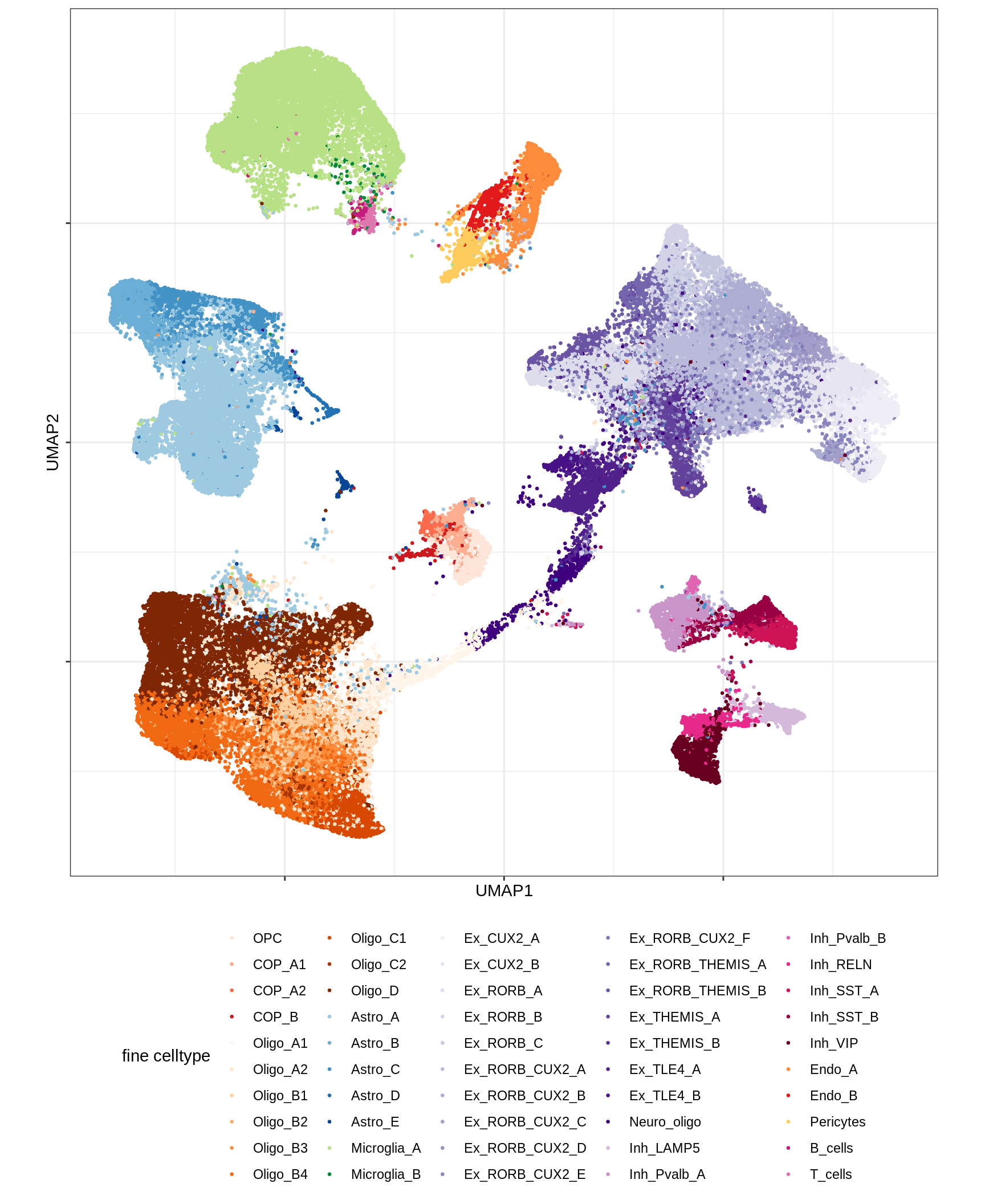
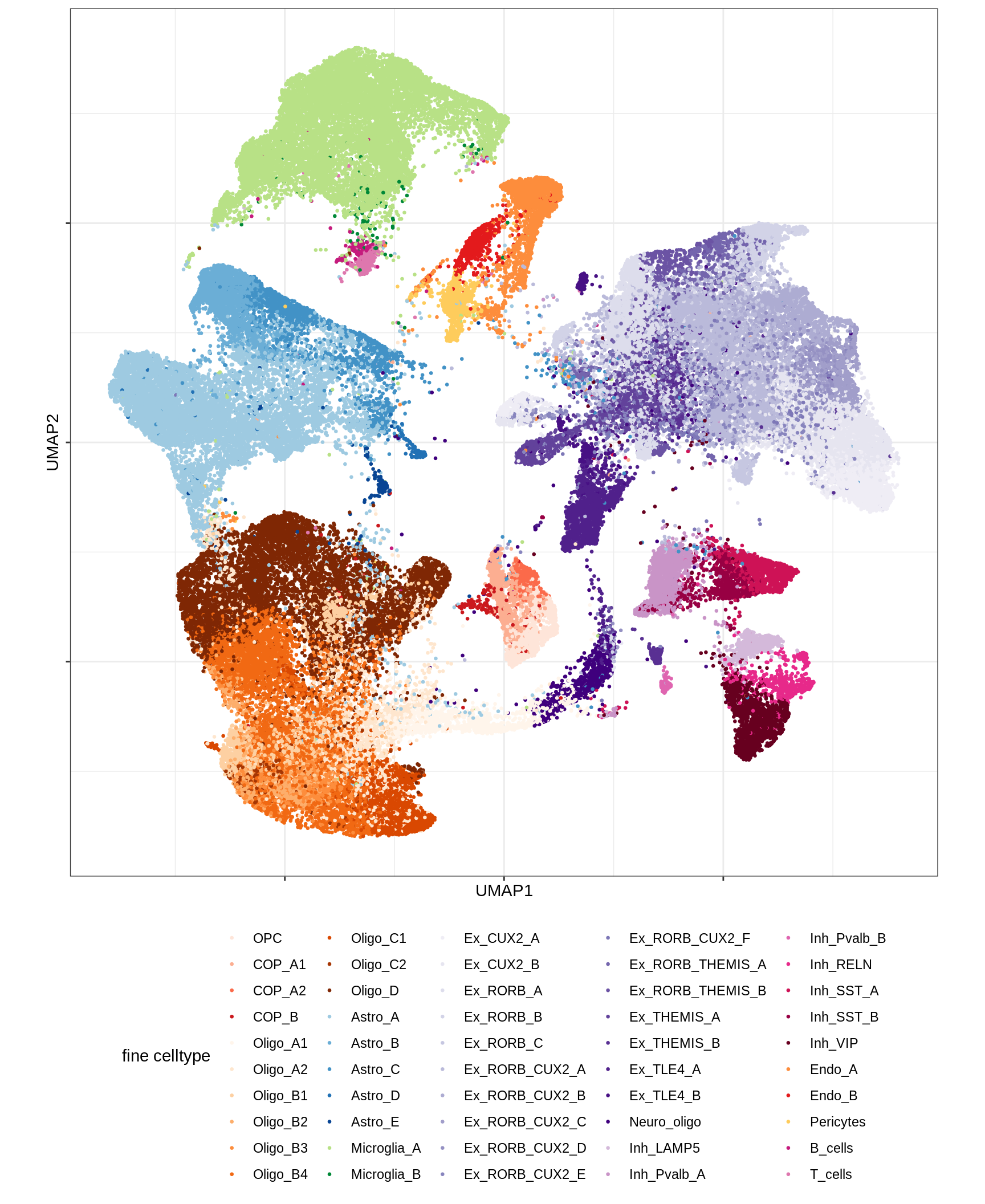
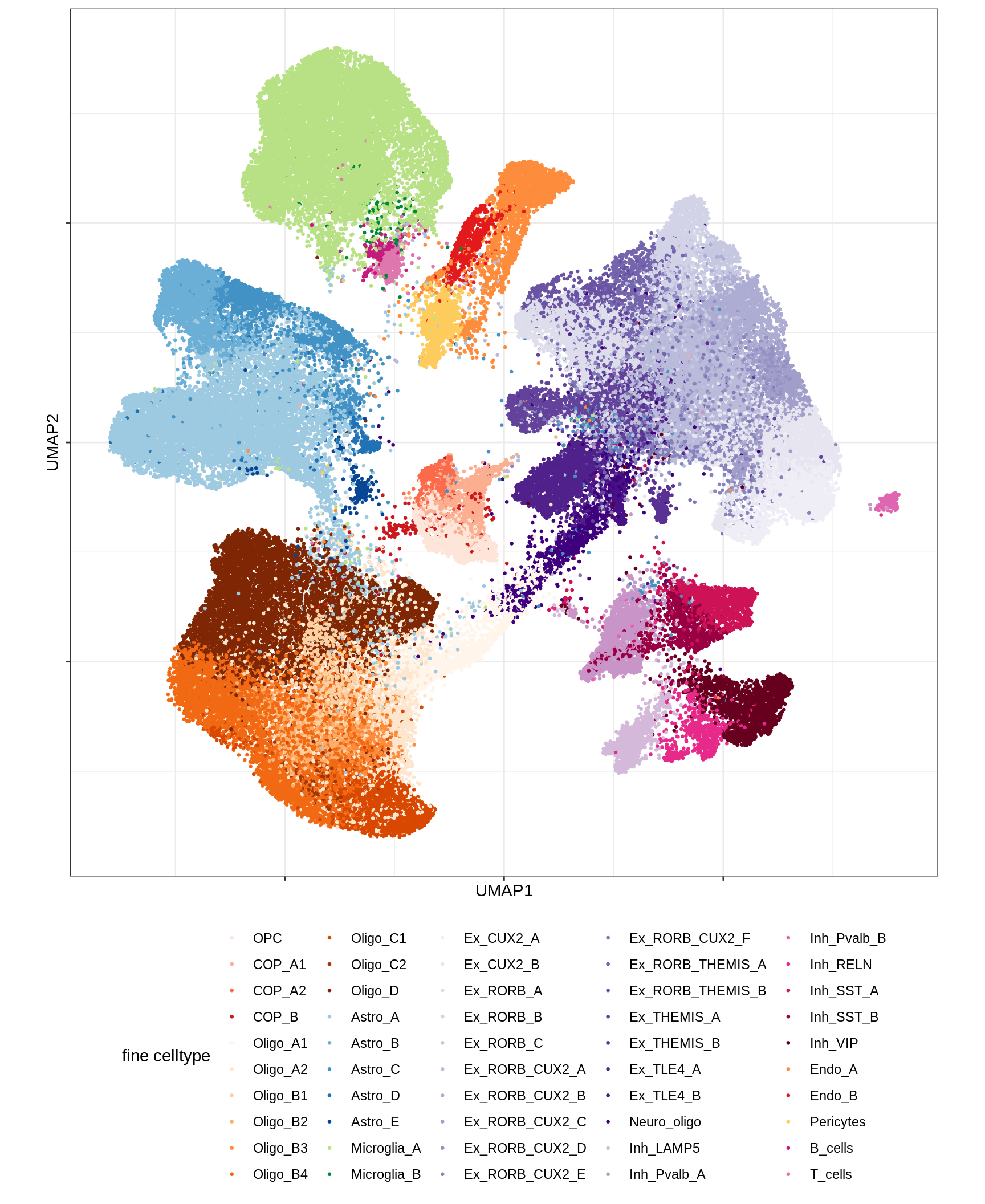
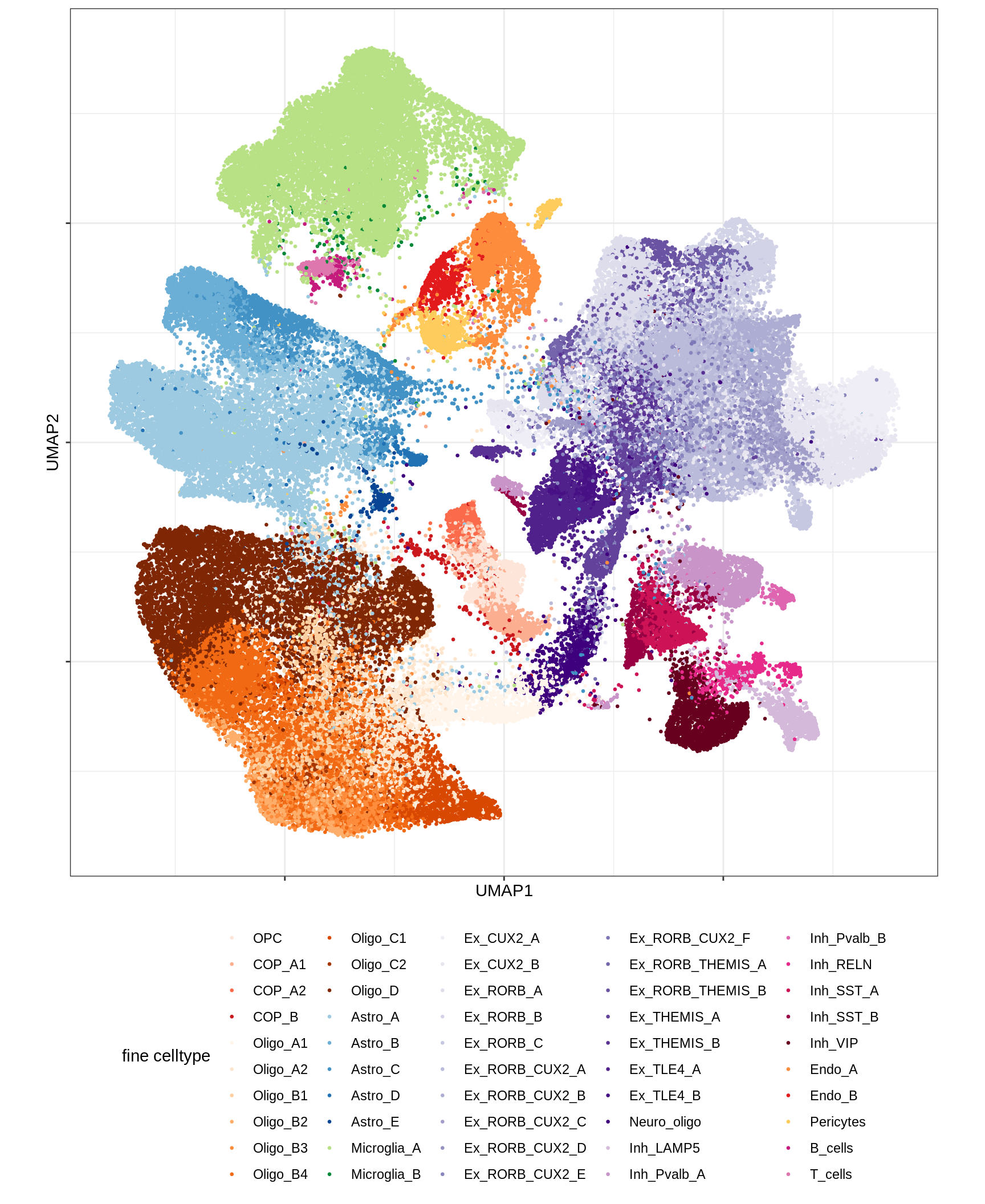
UMAP of final celltypes
for (d in unique(umap_dt$min_dist)) {
for (s in unique(umap_dt$spread)) {
cat(sprintf('### min_dist = %.2f, spread = %d\n', d, s))
print(plot_umap_final_celltypes(umap_dt[ min_dist == d & spread == s ],
final_cols))
cat('\n\n')
}
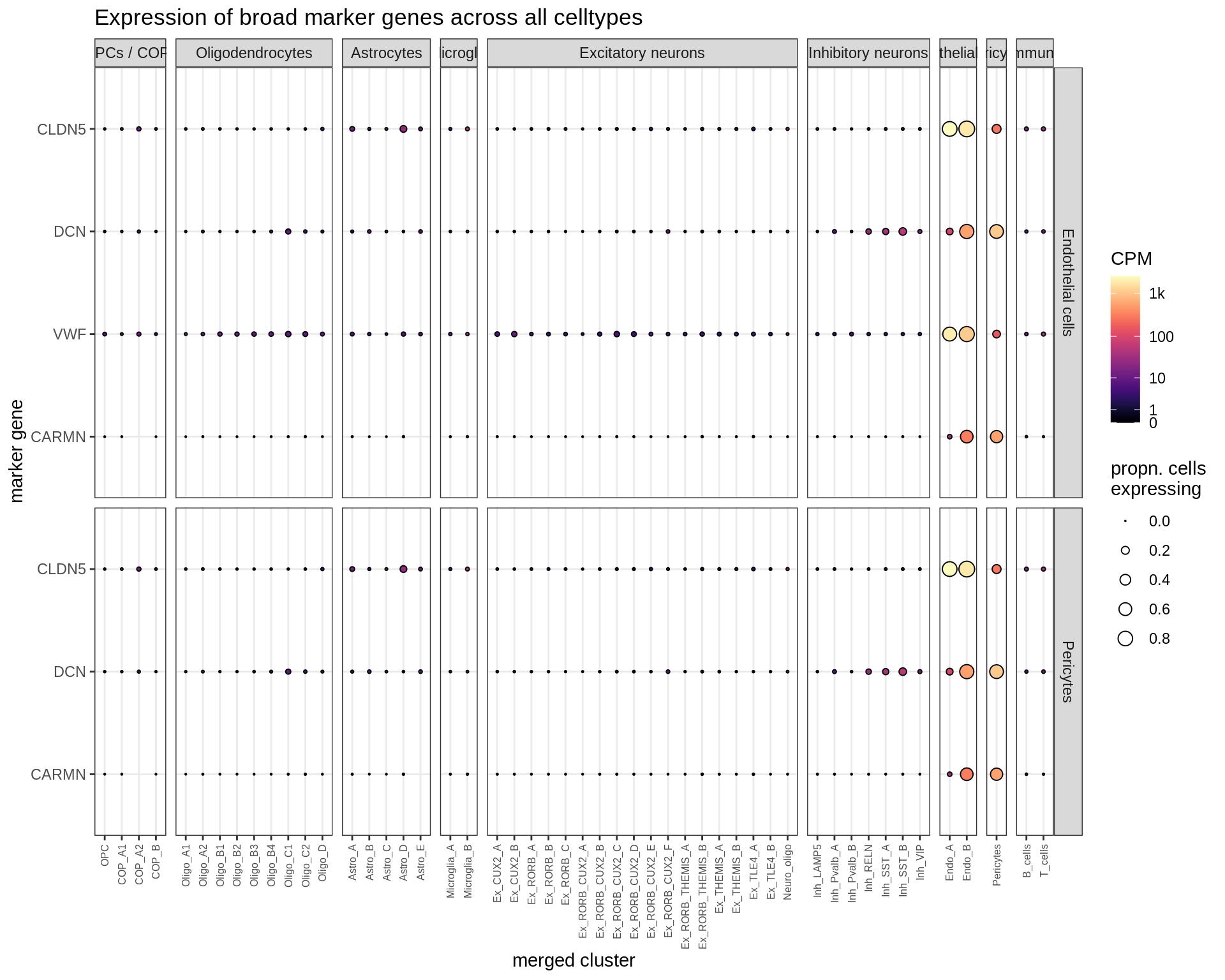
}Dotplots of hand-selected marker genes (broad)
(plot_dotplot(pb_fine, props_fine, byhand_ls$broad, labels_dt,
row_split = 'broad_class'))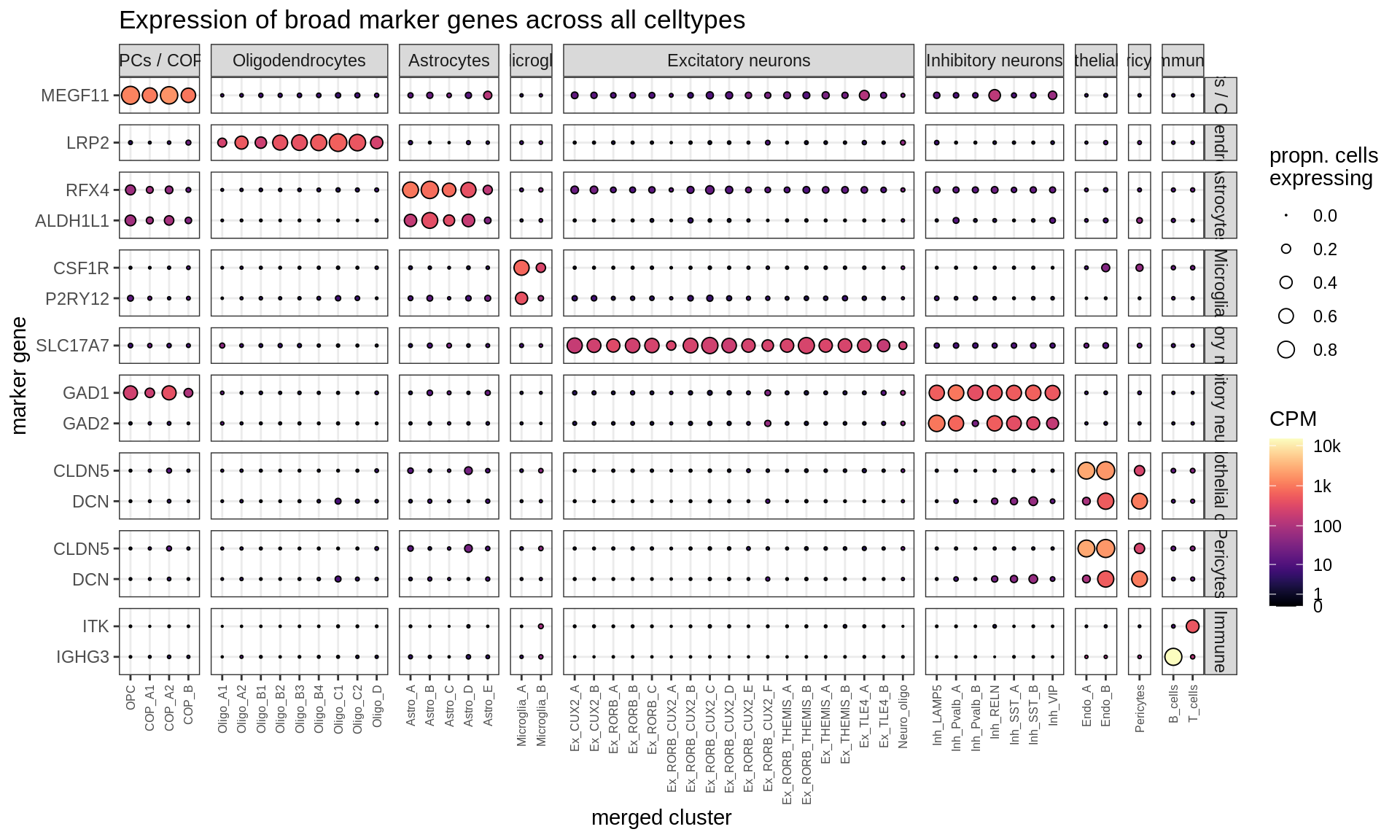
Dotplots of Dheeraj’s marker genes, compact (broad)
remove_dt = list(
`OPCs / COPs` = c("VCAN"),
Oligodendrocytes = c("GADD45B", "MOBP", "FSTL5"),
Astrocytes = c("ALDH1L1", "LRAT", "WIF1", "LINC00499"),
Microglia = c("CD74", "APOE"),
`Excitatory neurons` = c("ST18", "SV2B"),
`Inhibitory neurons` = c("CUX2", "GAD2", "EYA4"),
`Endothelial cells` = c("VWF"),
Immune = c("IGH4", "CD96")
) %>% imap( ~ data.table( broad_class = .y, symbol = .x, remove = TRUE )) %>%
rbindlist
dheeraj_tweak = merge(dheeraj_dt, remove_dt, by = c('broad_class', 'symbol'),
all.x = TRUE) %>%
.[ is.na(remove), remove := FALSE ] %>%
.[ remove == FALSE ] %>%
.[, broad_class := factor(broad_class, levels = broad_ord) %>%
fct_collapse(Endo = c("Endothelial cells", "Pericytes")) %>%
fct_recode(
OPCs = 'OPCs / COPs', Oligos = "Oligodendrocytes",
Micro = 'Microglia', Astros = 'Astrocytes',
`Inhib. neurons` = 'Inhibitory neurons', Imm = 'Immune'
)] %>%
.[, symbol := factor(symbol, levels = unique(dheeraj_dt$symbol) )] %>%
unique %>%
.[ order(broad_class, symbol) ] %>%
.[, symbol := as.character(symbol)]
labels_tweak = copy(labels_dt) %>%
.[, type_broad := fct_collapse(type_broad,
Endo = c("Endothelial cells", "Pericytes")) %>%
fct_recode(
OPCs = 'OPCs / COPs', Oligos = "Oligodendrocytes",
Micro = 'Microglia', Astros = 'Astrocytes',
`Inhib. neurons` = 'Inhibitory neurons', Imm = 'Immune'
) ]
(plot_dotplot(pb_fine, props_fine, dheeraj_tweak, labels_tweak,
row_split = 'broad_class'))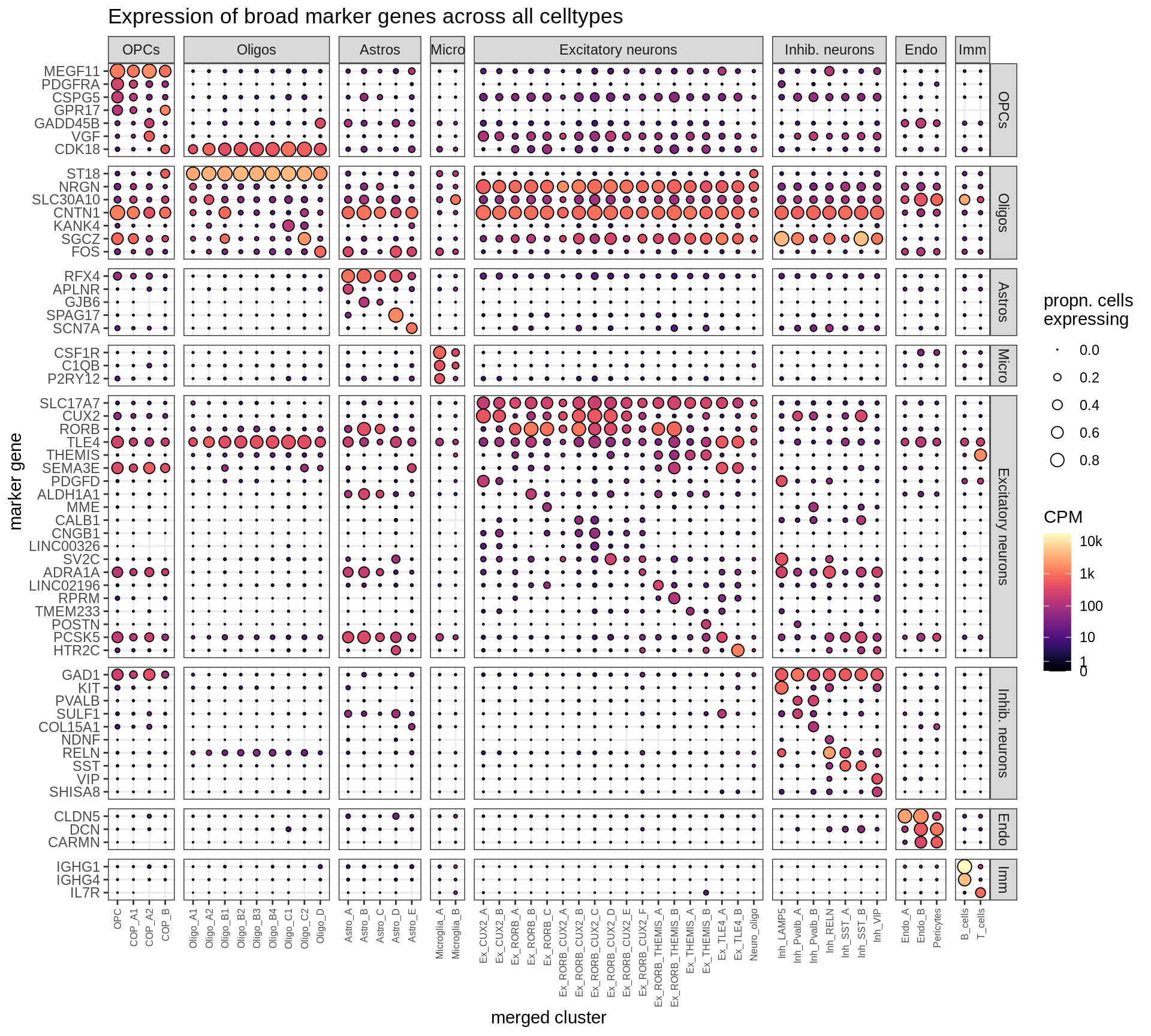
Dotplots of Dheeraj’s marker genes, all (broad)
(plot_dotplot(pb_fine, props_fine, dheeraj_dt, labels_dt,
row_split = 'broad_class'))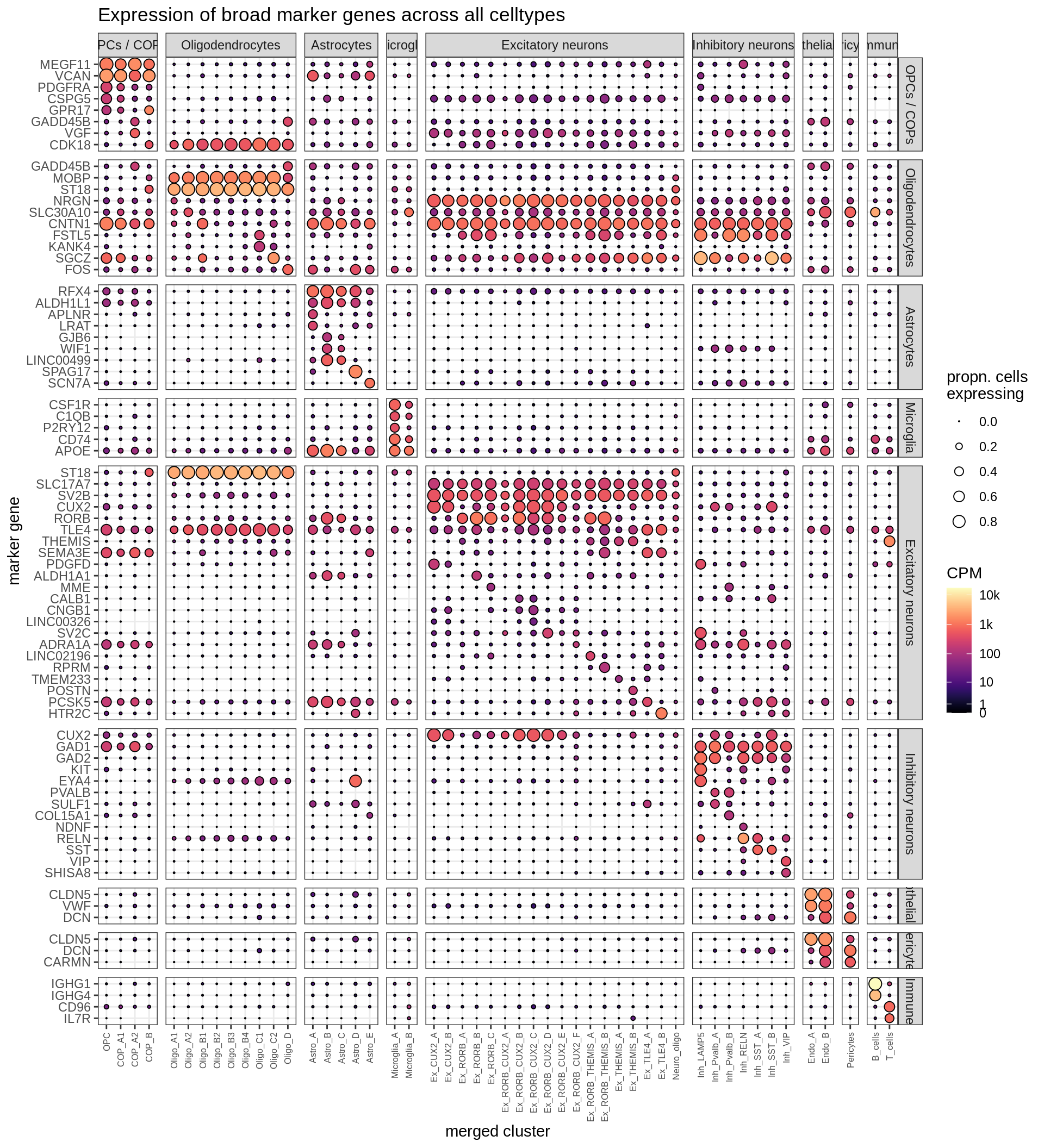
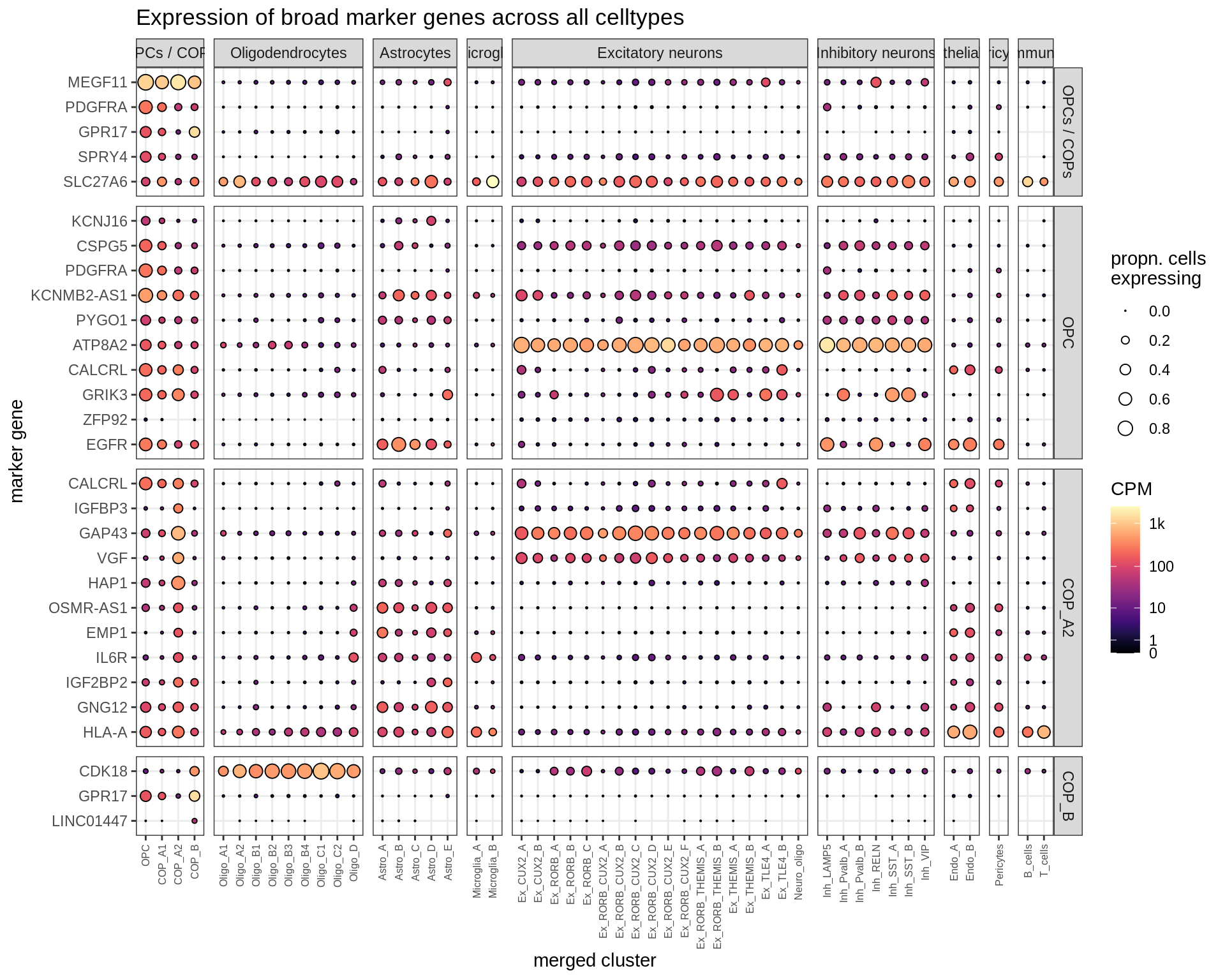
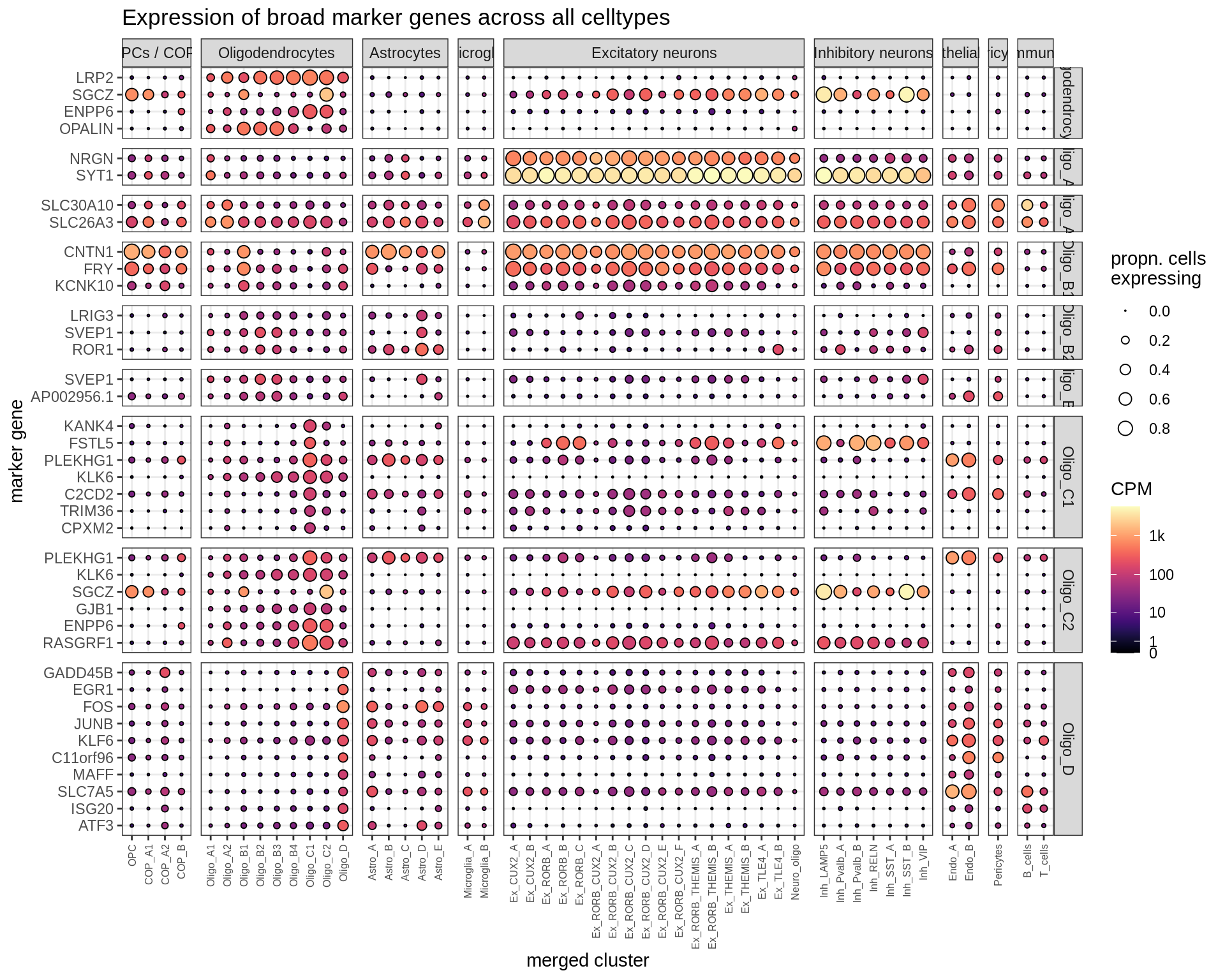
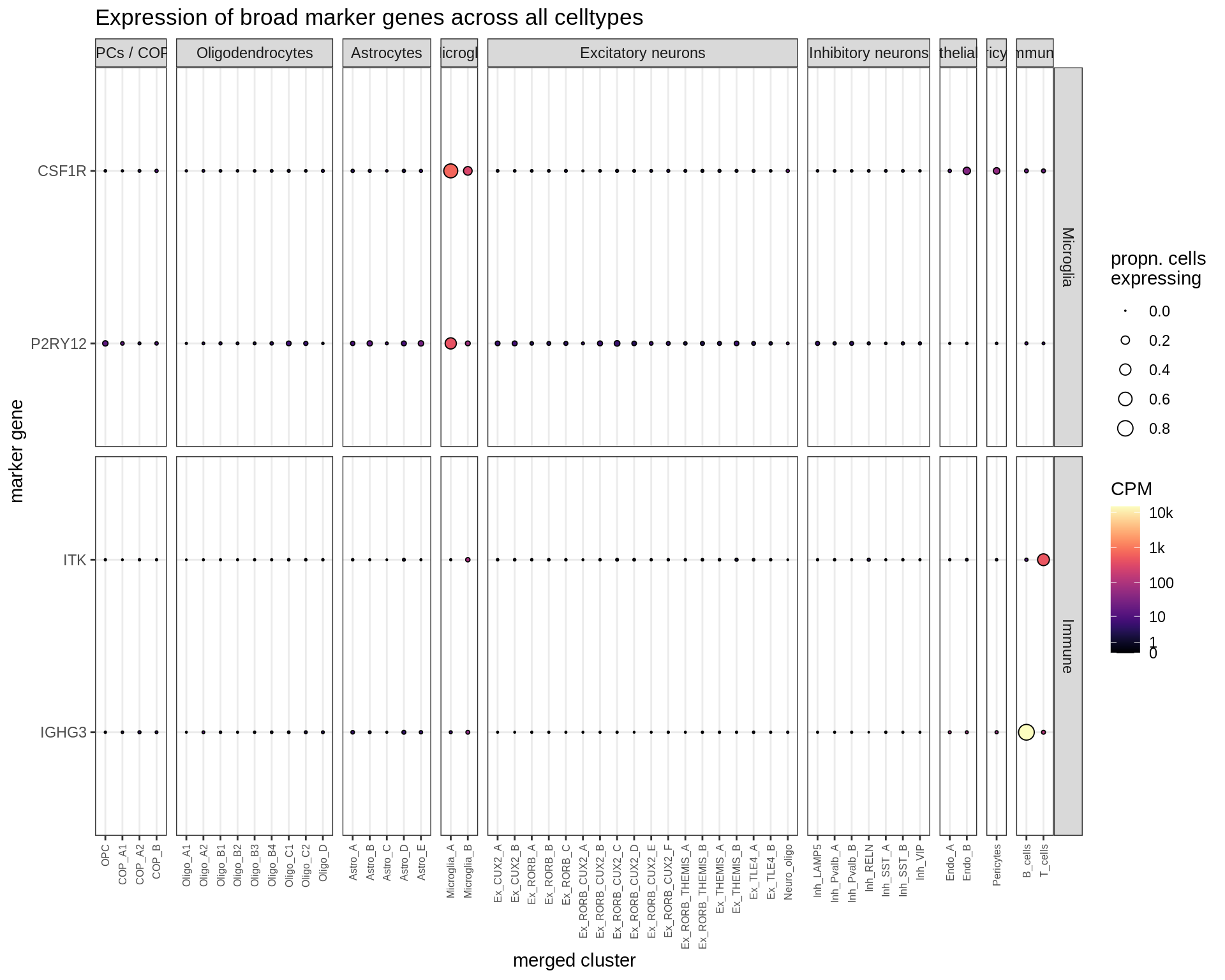
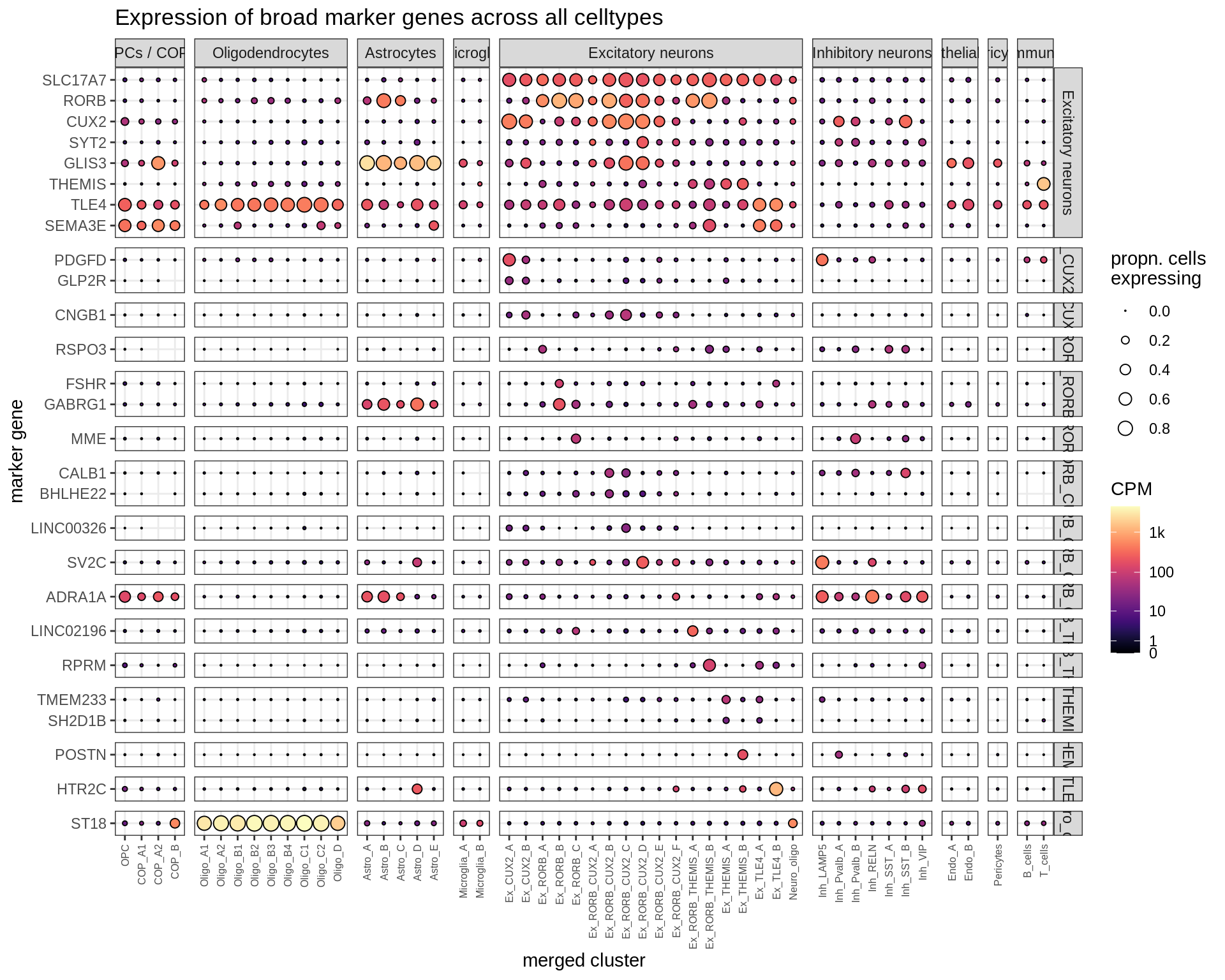
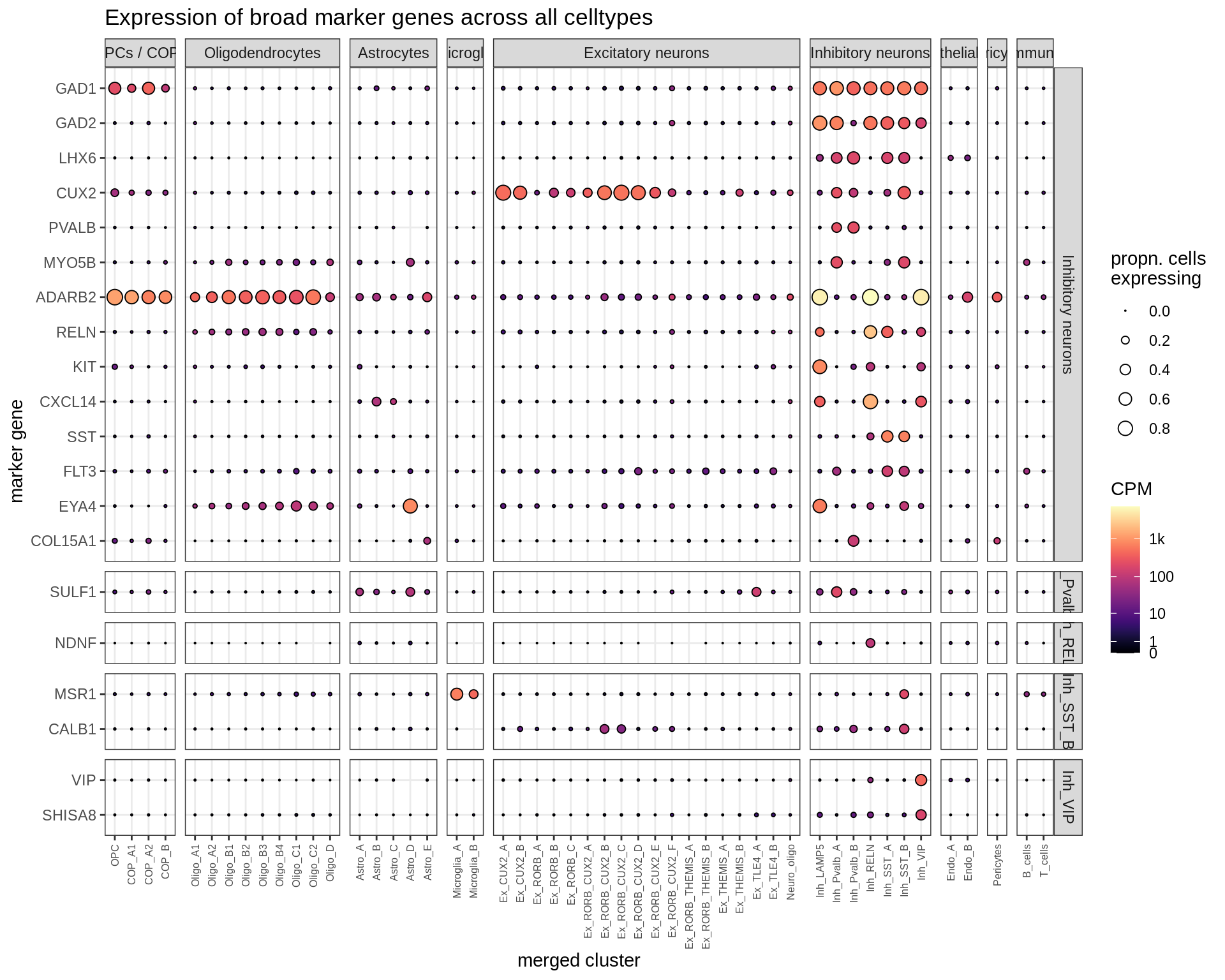
Dotplots of hand-selected marker genes (split by cluster)
for (b in names(broad_list)) {
cat('### ', b, '\n')
byhand_dt = rbind(
byhand_ls$broad[broad_class %in% broad_list[[b]],
.(fine_class = broad_class, symbol)],
byhand_ls$fine[broad_class %in% broad_list[[b]],
.(fine_class, symbol)]
) %>%
.[, fine_class := fine_class %>% fct_drop %>% fct_relevel(broad_list[[b]])]
suppressWarnings(print(plot_dotplot(pb_fine, props_fine, byhand_dt, labels_dt,
row_split = 'fine_class')))
cat('\n\n')
}OPCs / COPs
Oligodendrocytes
Astrocytes
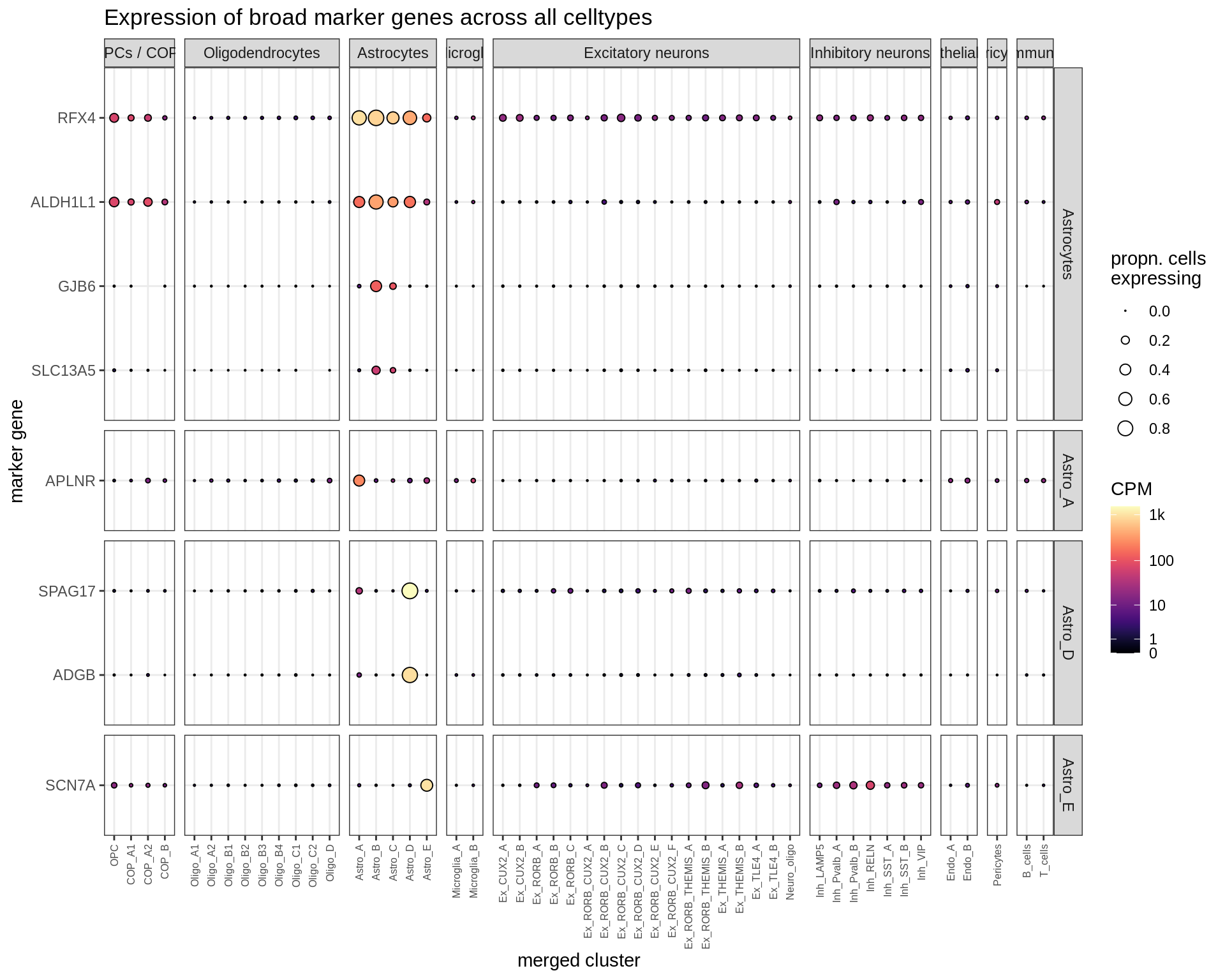
Microglia + Immune
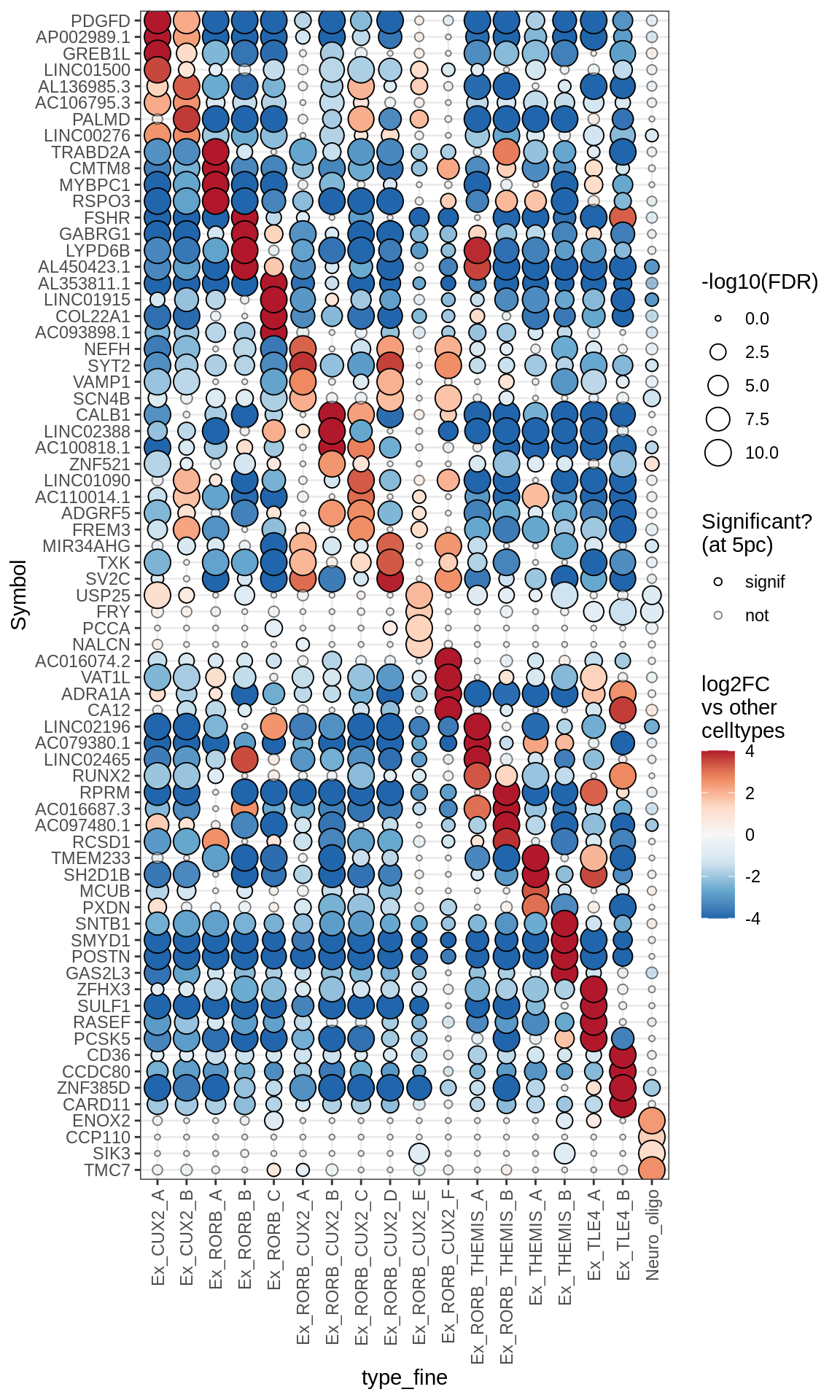
Excitatory neurons
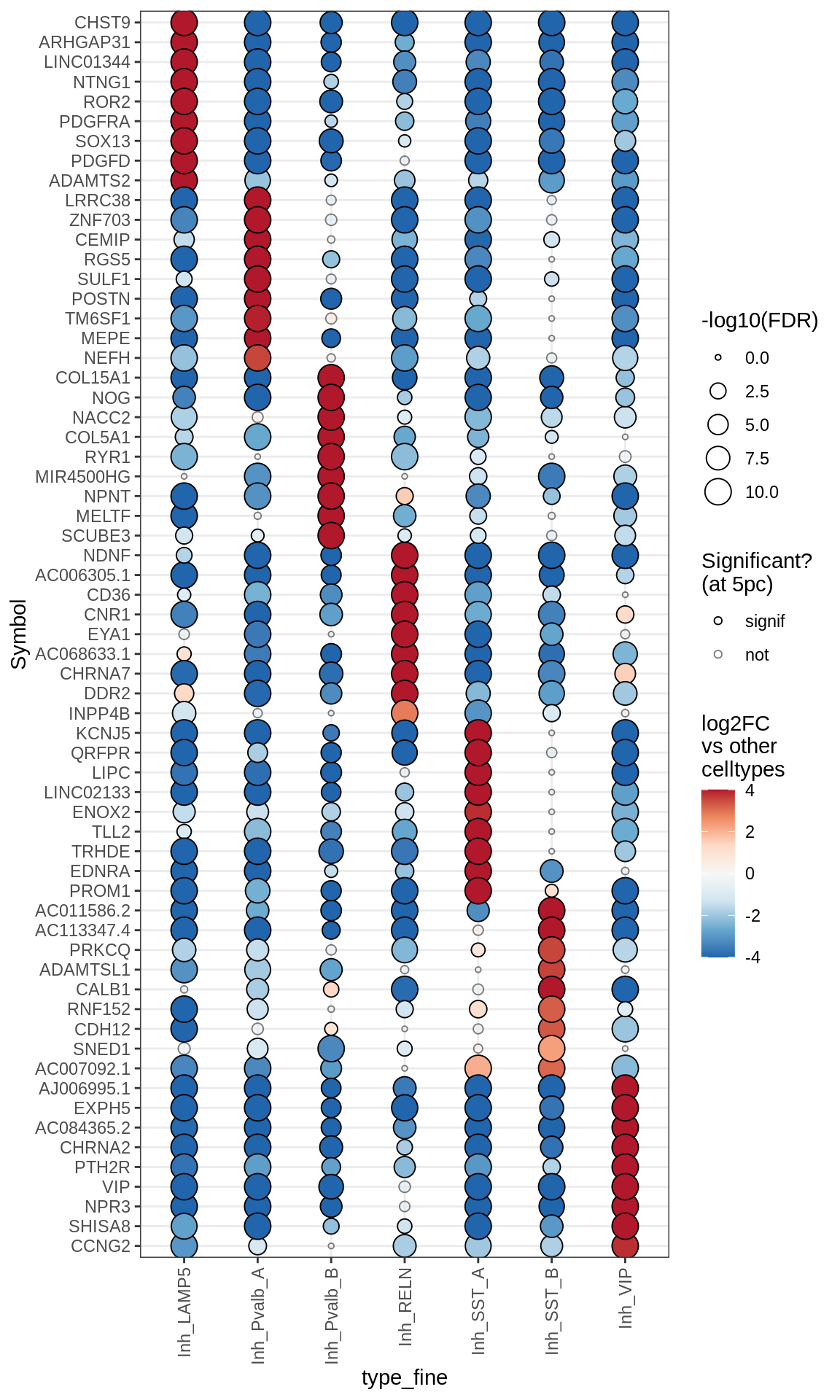
Inhibitory neurons
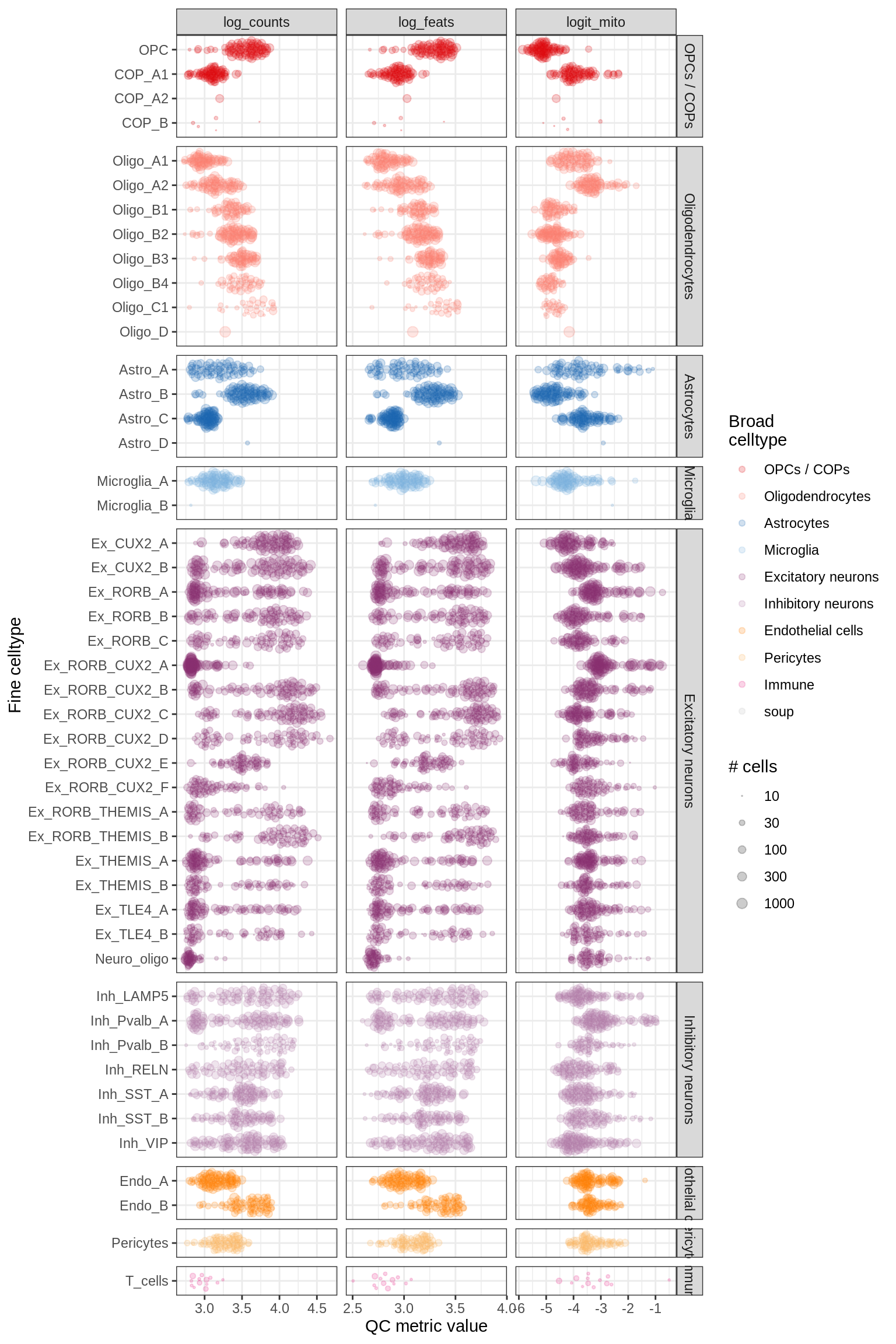
QC at pseudobulk level
for (m in c('WM', 'GM')) {
cat('### ', m, '\n')
print(plot_pb_qc(pb_qc_dt[ matter == m ], min_cells = min_cells))
cat('\n\n')
}WM
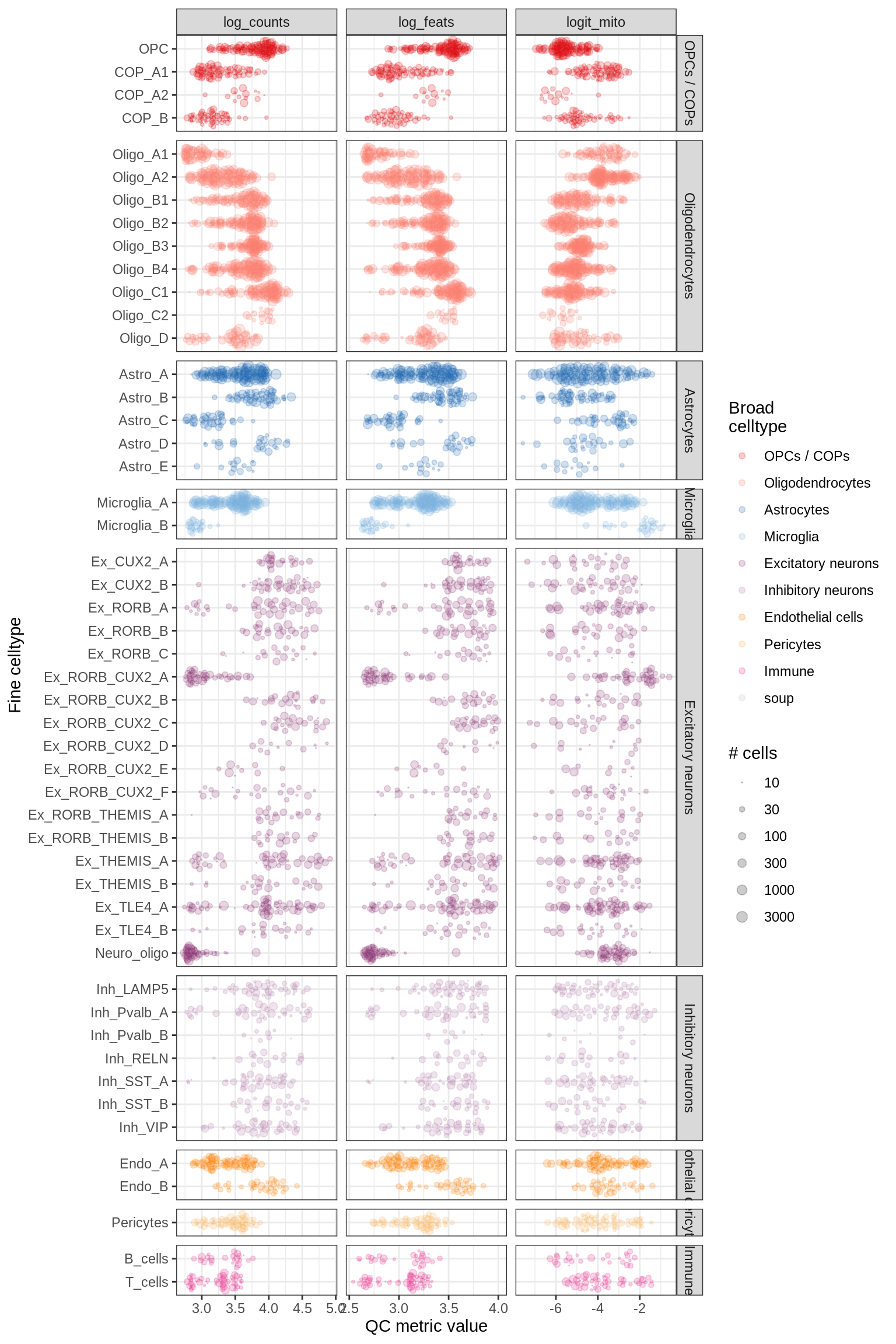
GM
Dotplots of muscat results (all samples, broad)
for (b in names(broad_list)) {
cat('### ', b, '\n')
print(plot_dotplot_muscat(muscat_all, labels_dt, broad_list[[b]],
fdr_cut = 0.05))
cat('\n\n')
}OPCs / COPs
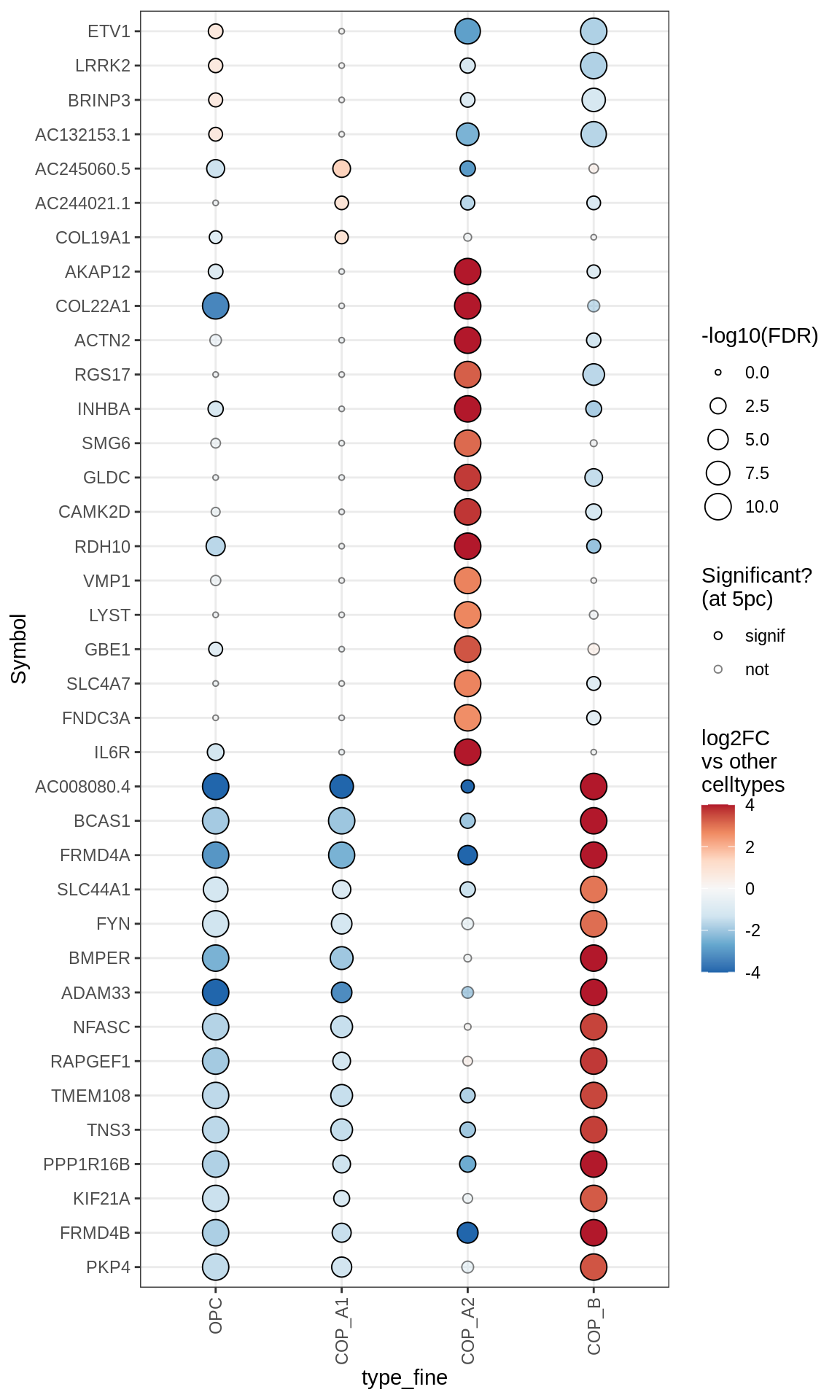
Oligodendrocytes
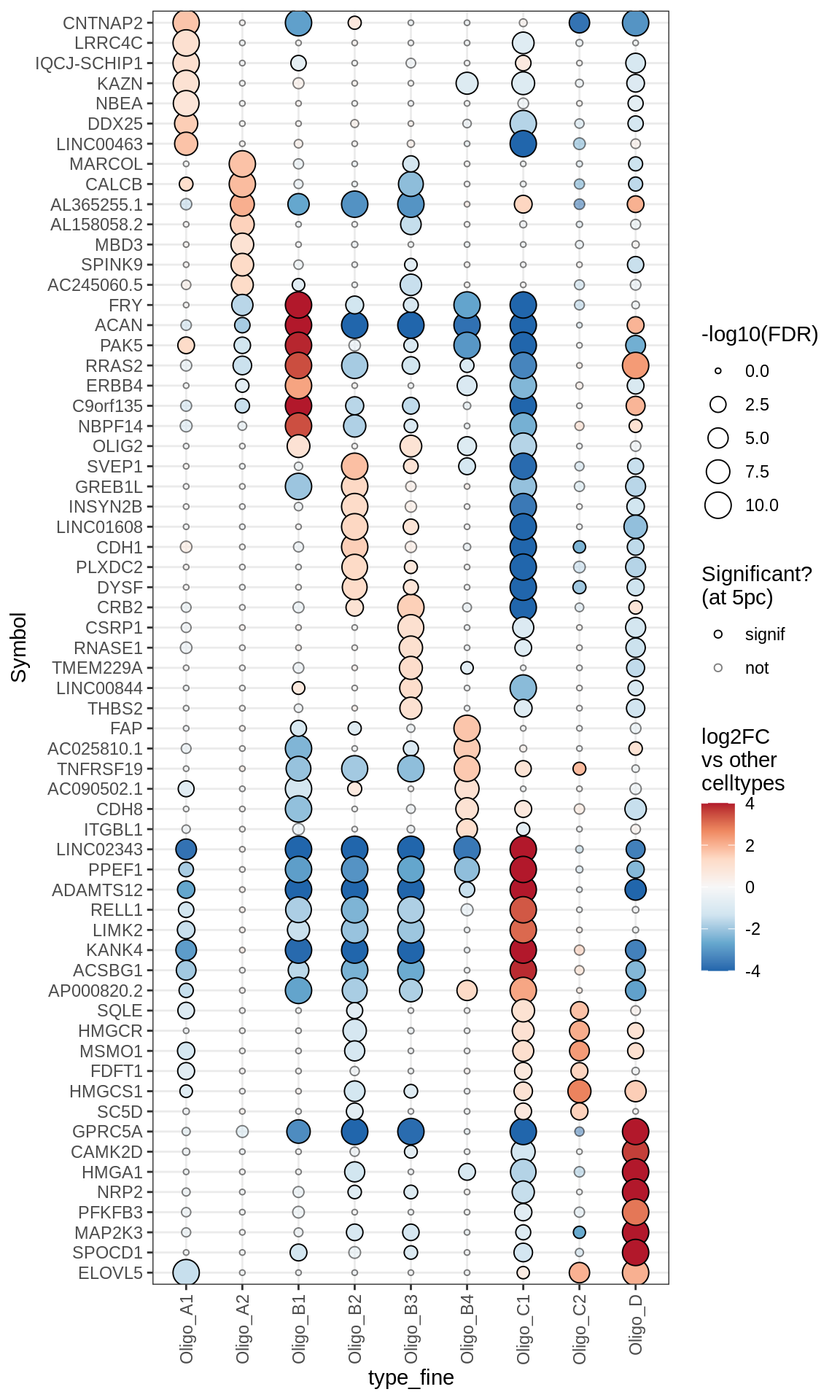
Astrocytes
Microglia + Immune
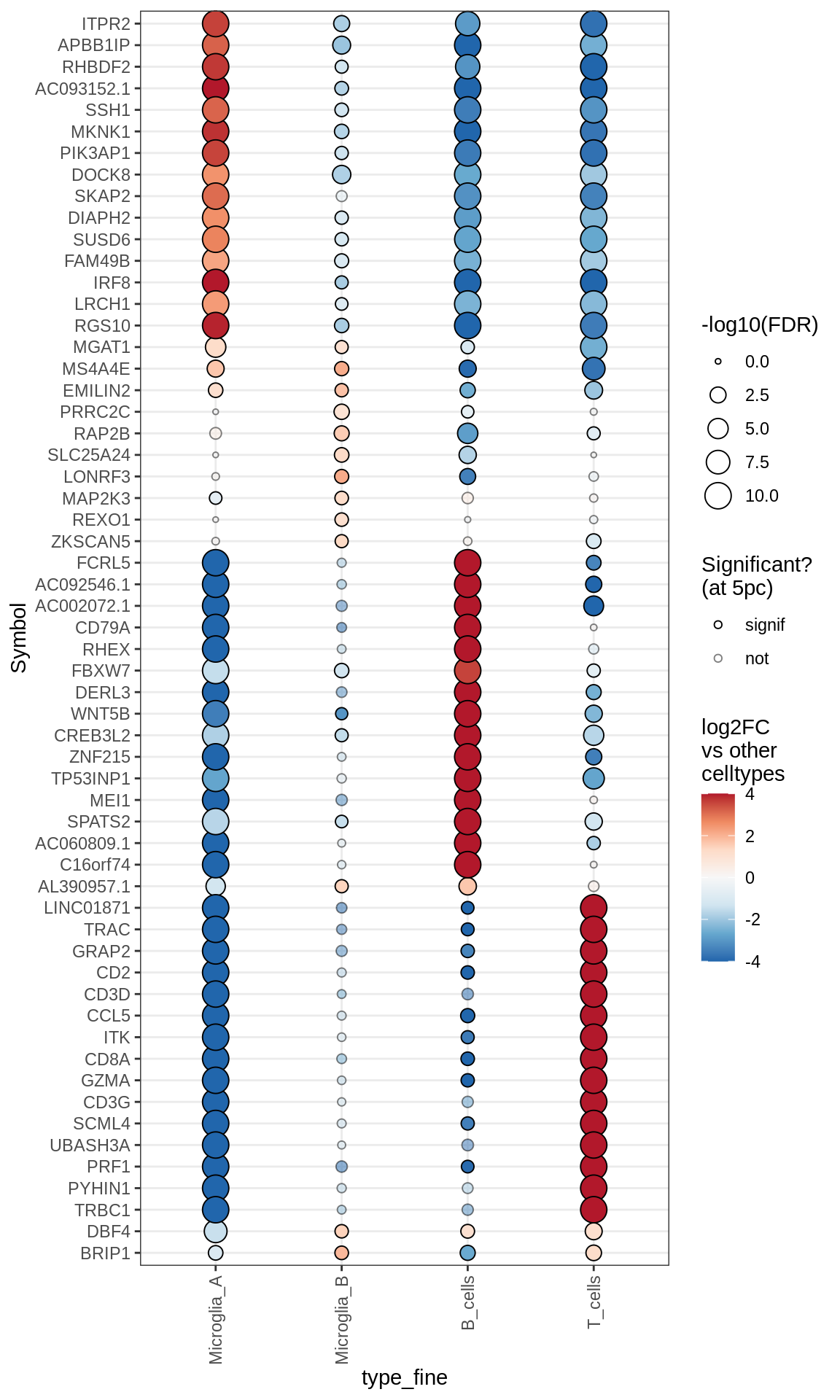
Excitatory neurons
Inhibitory neurons
Endothelial + Pericytes
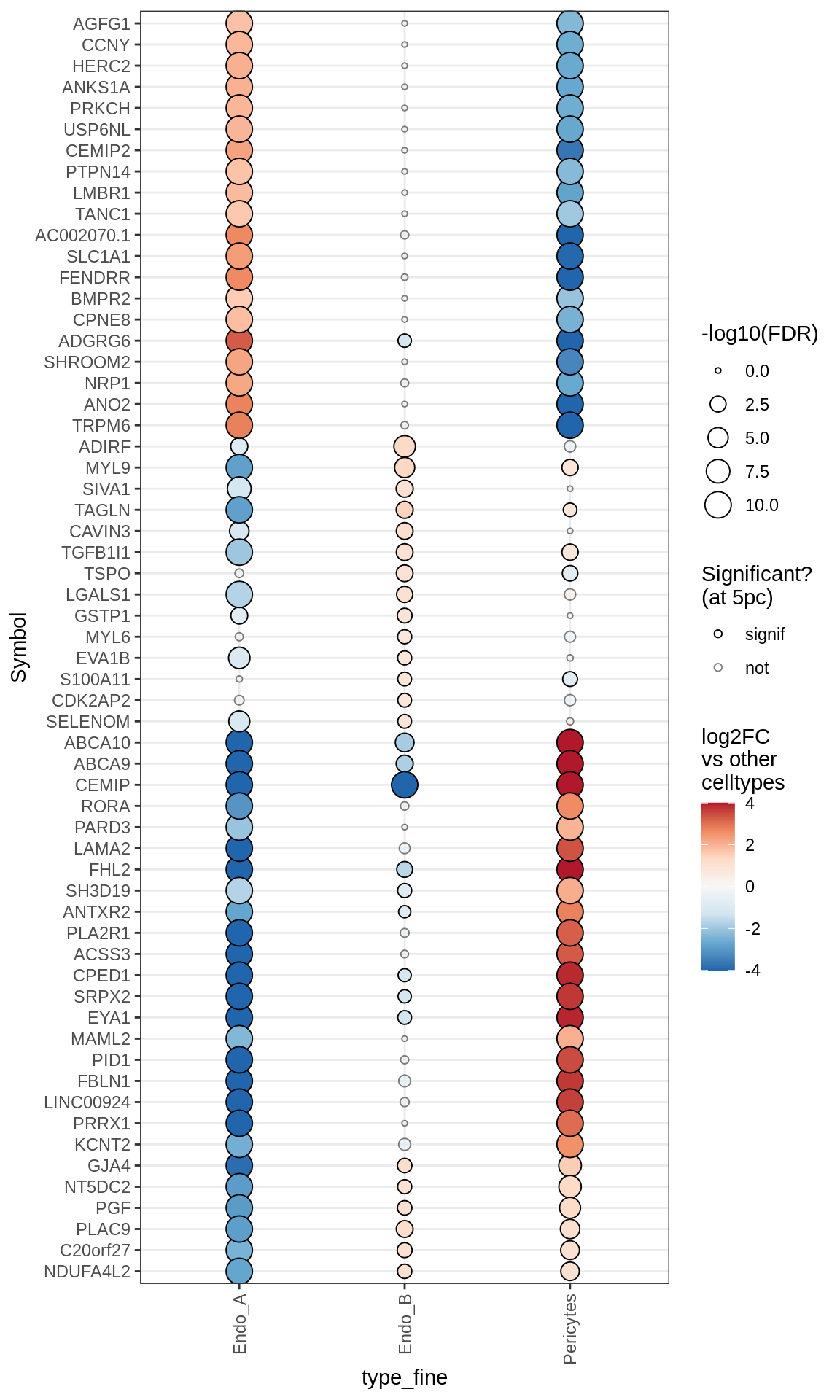
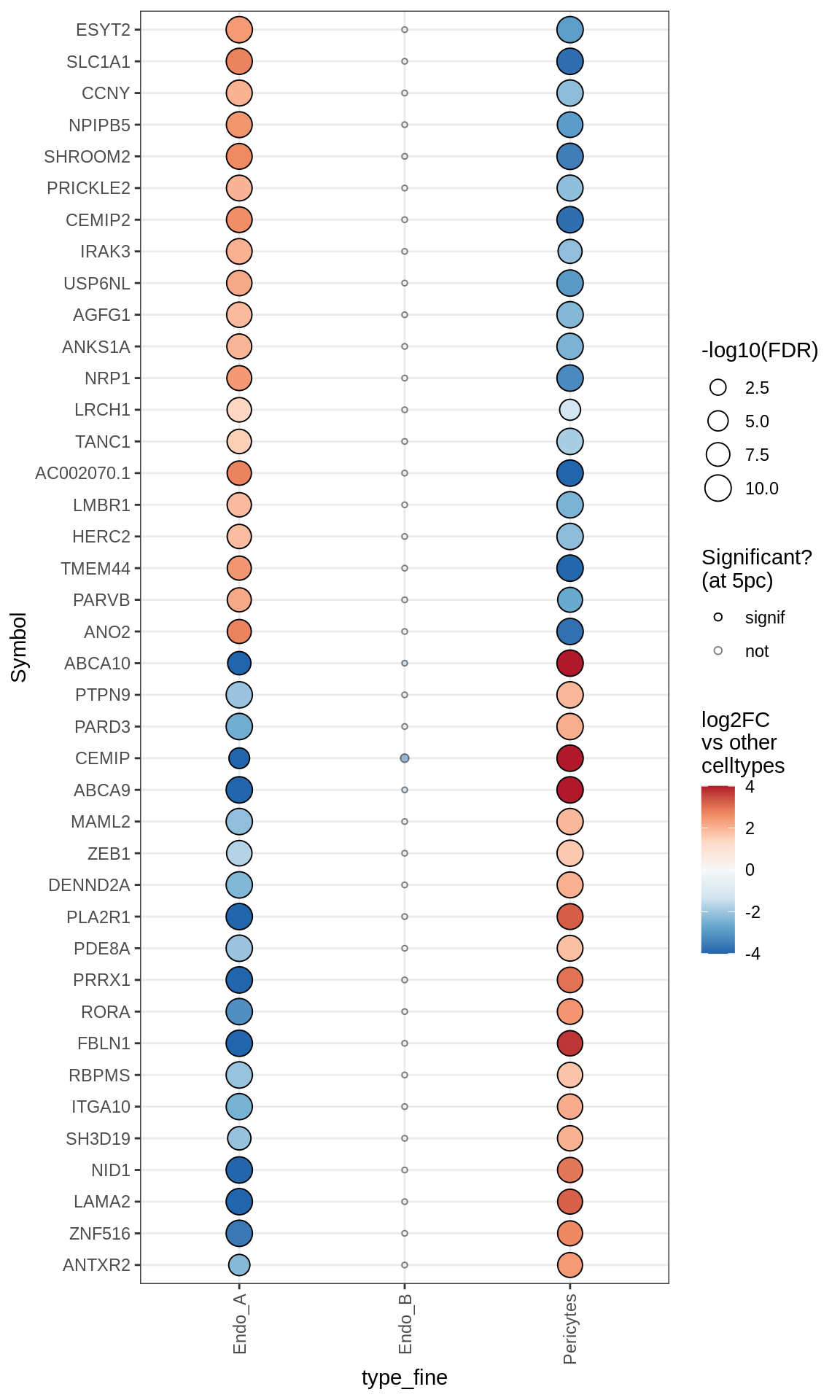
Dotplots of muscat results (healthy samples, broad)
for (b in names(broad_list)) {
cat('### ', b, '\n')
print(plot_dotplot_muscat(muscat_ctrl, labels_dt, broad_list[[b]],
fdr_cut = 0.05))
cat('\n\n')
}OPCs / COPs
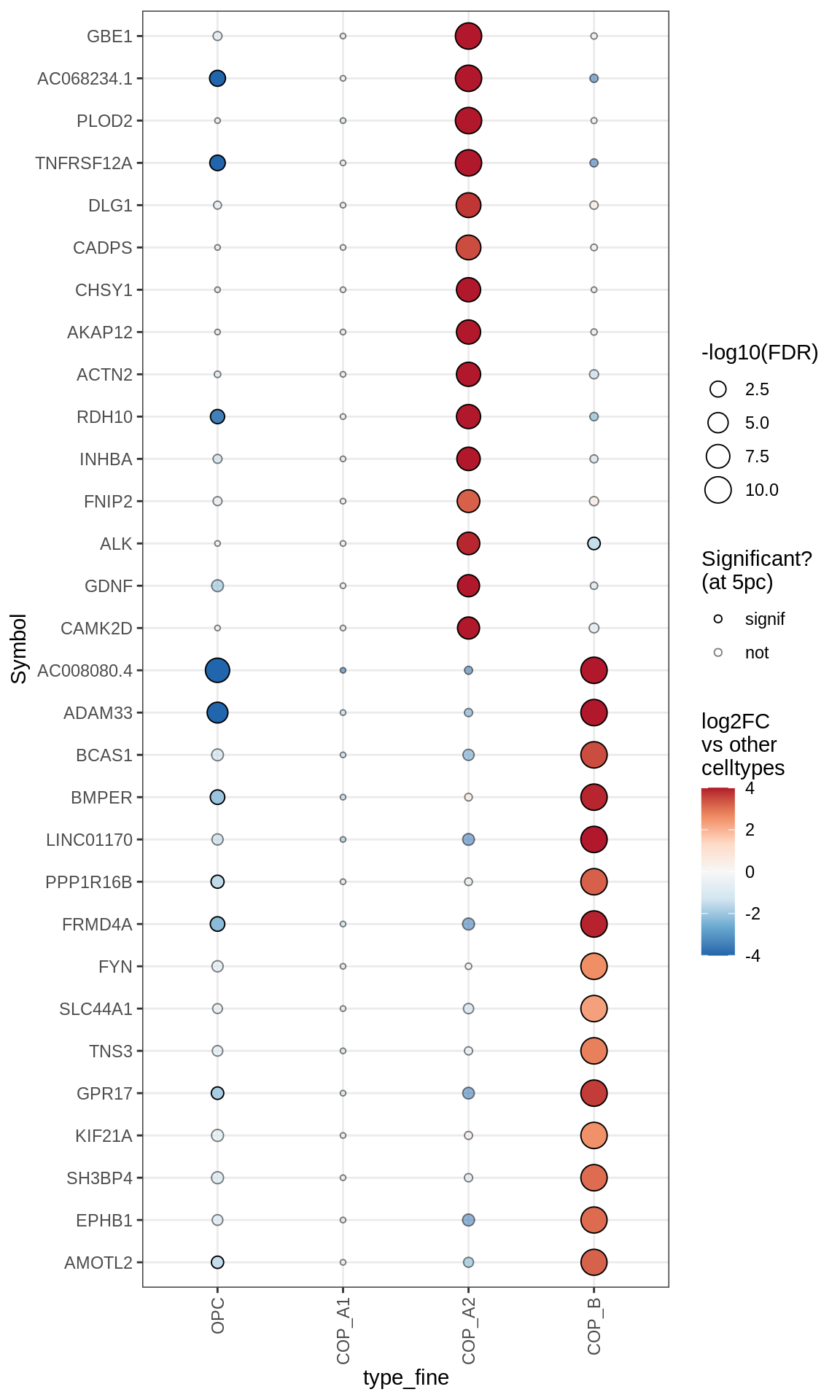
Oligodendrocytes
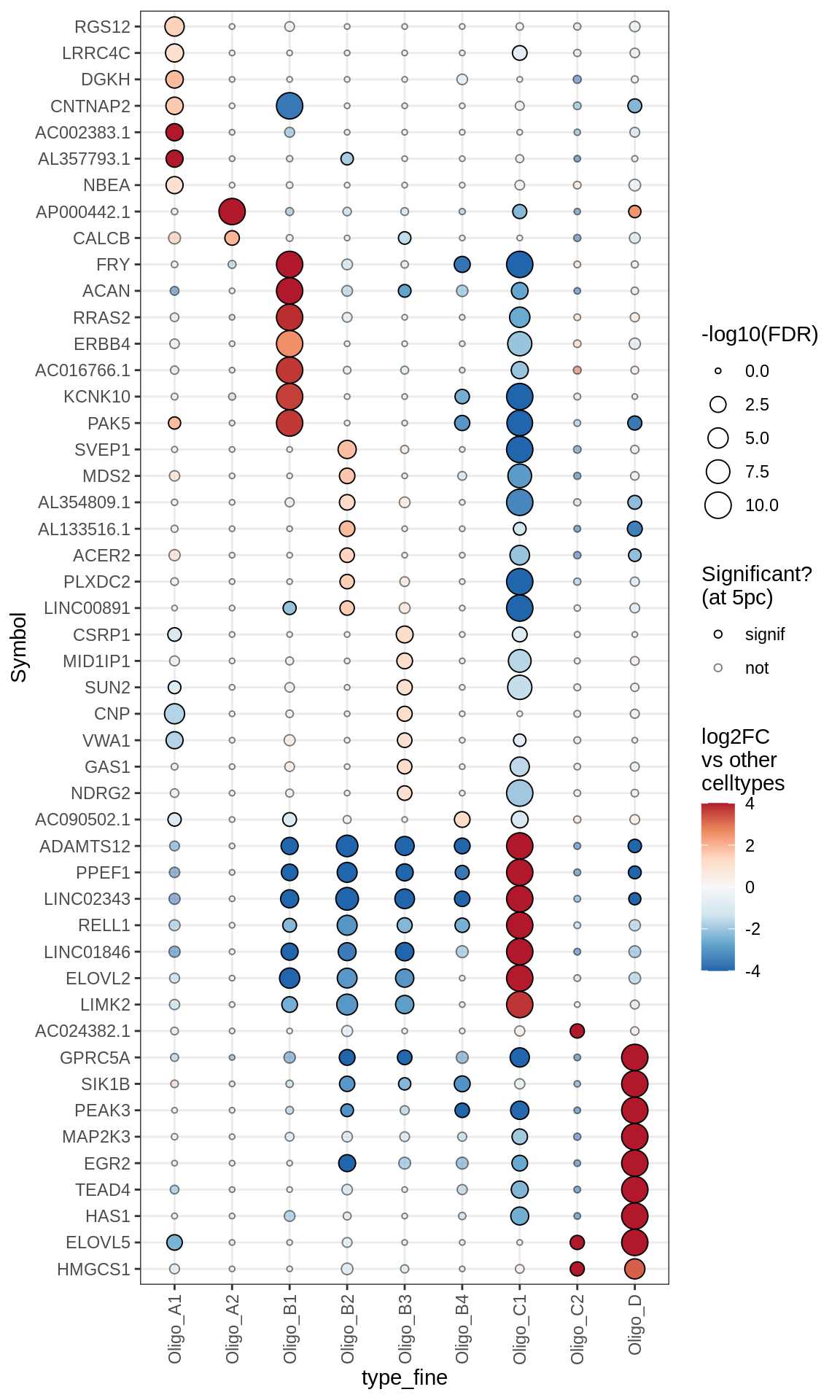
Astrocytes
Microglia + Immune
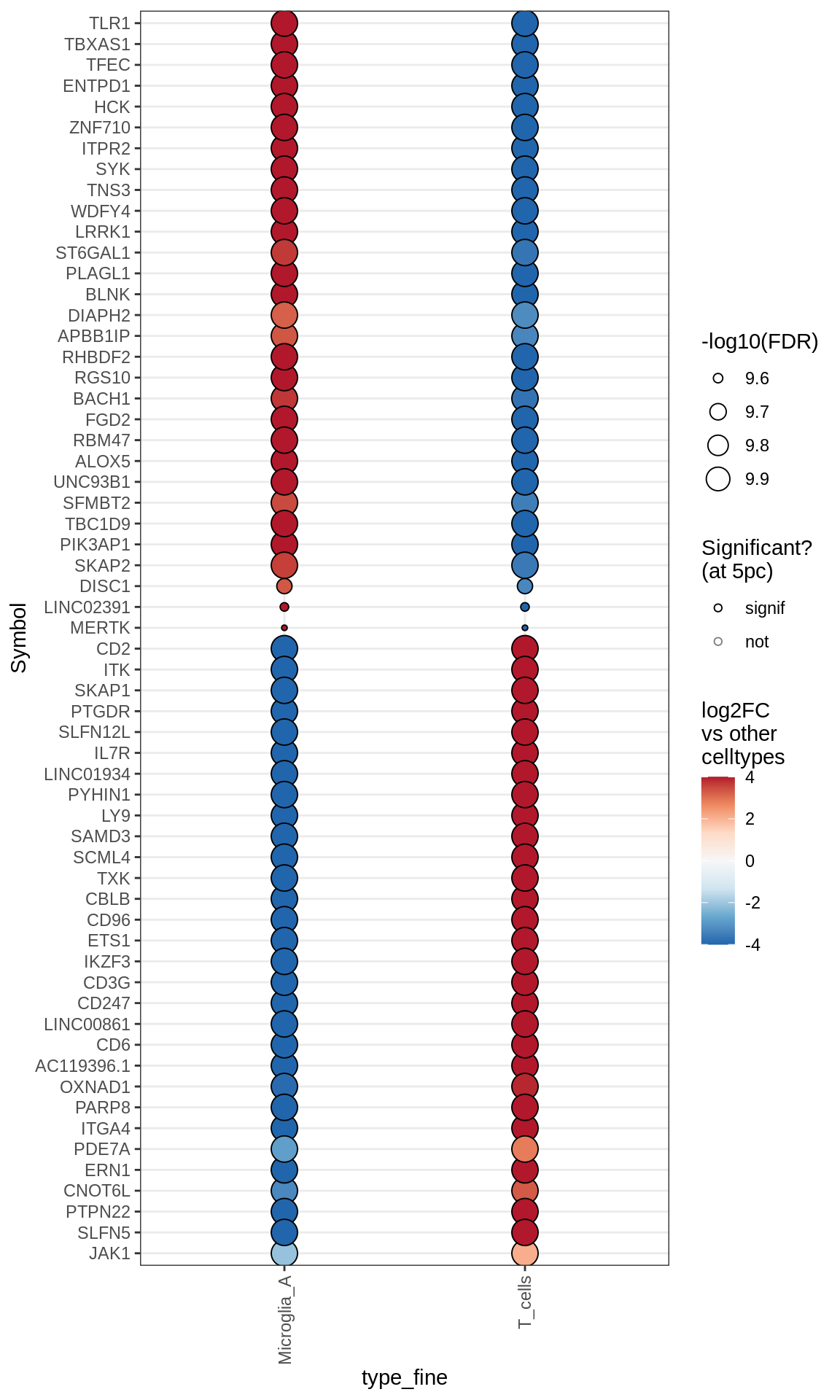
Excitatory neurons
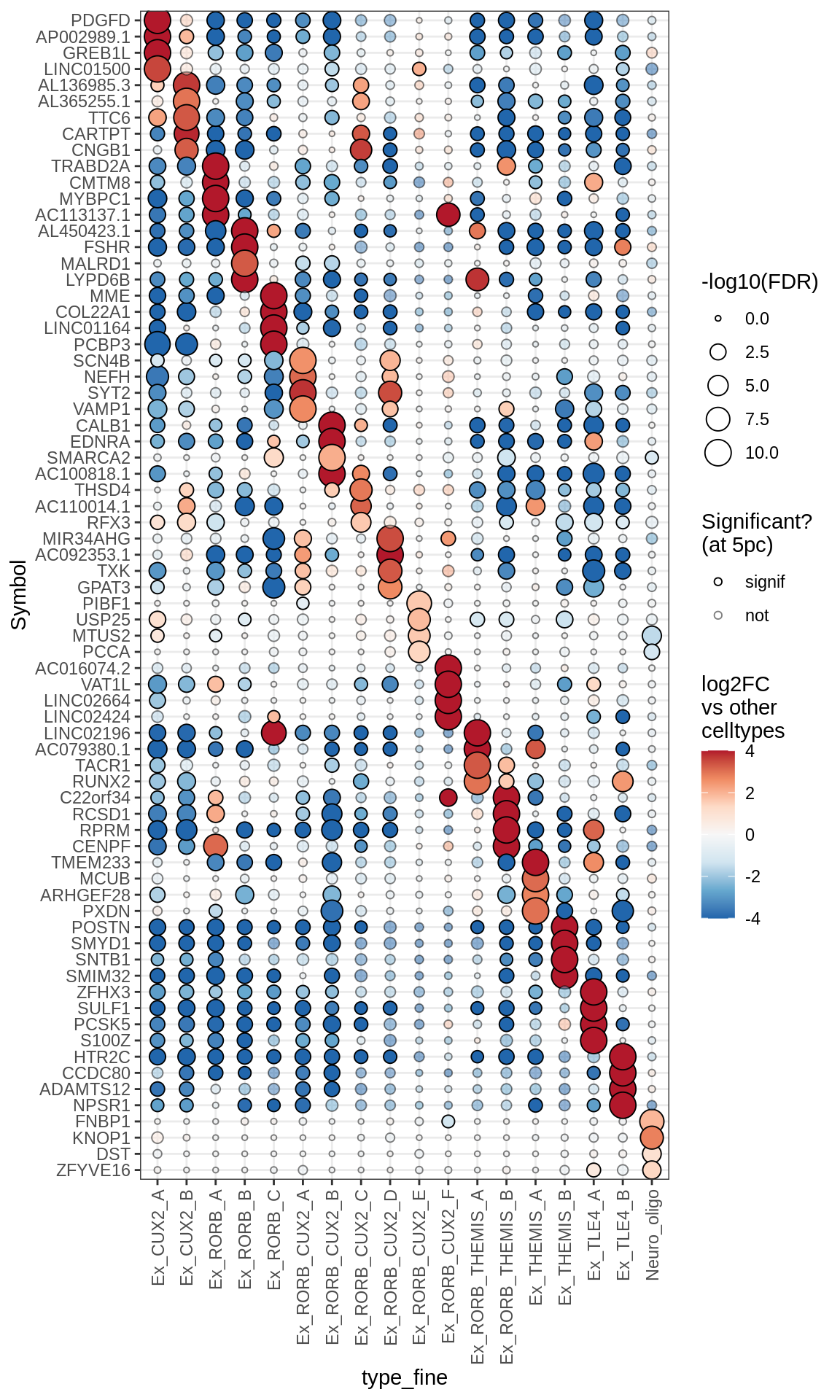
Inhibitory neurons
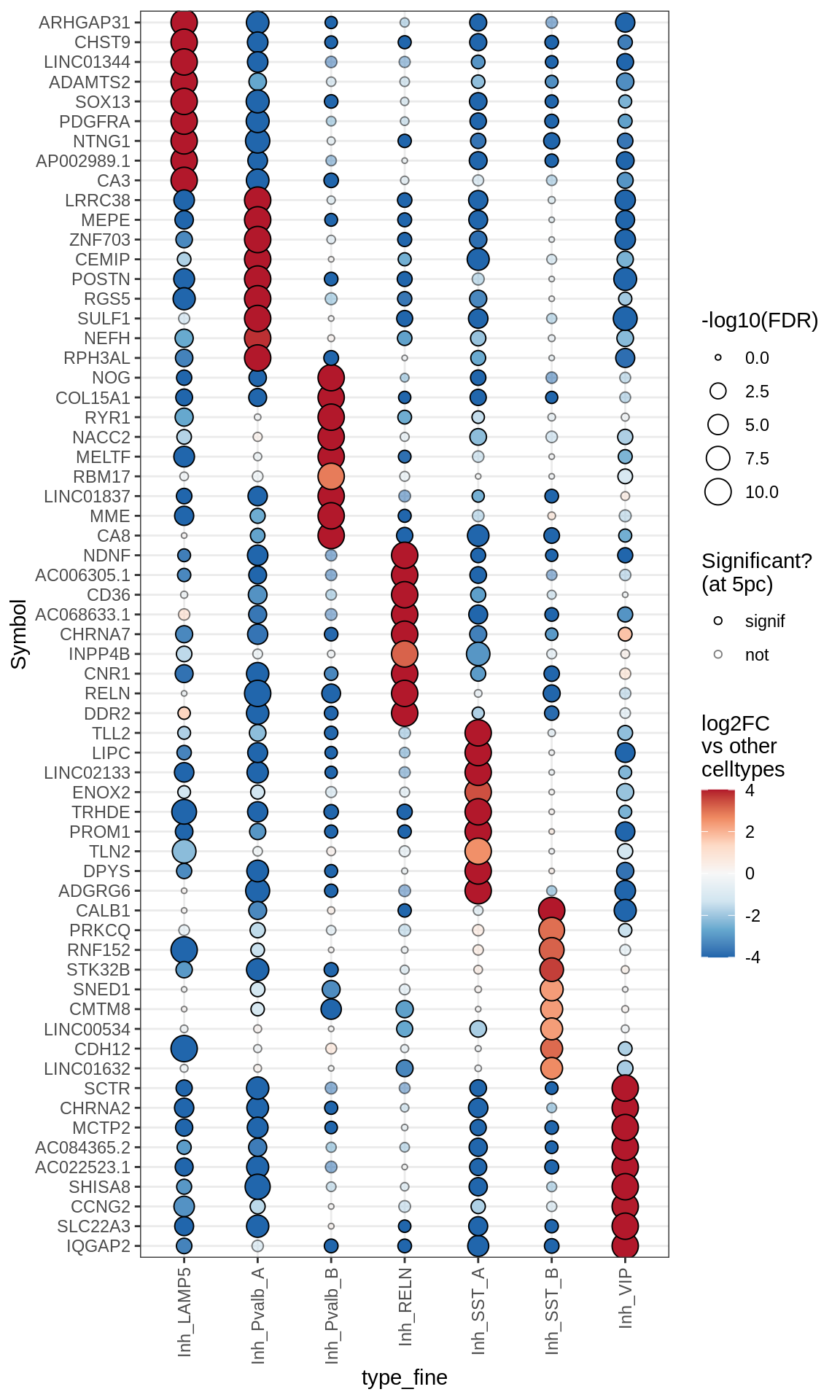
Endothelial + Pericytes
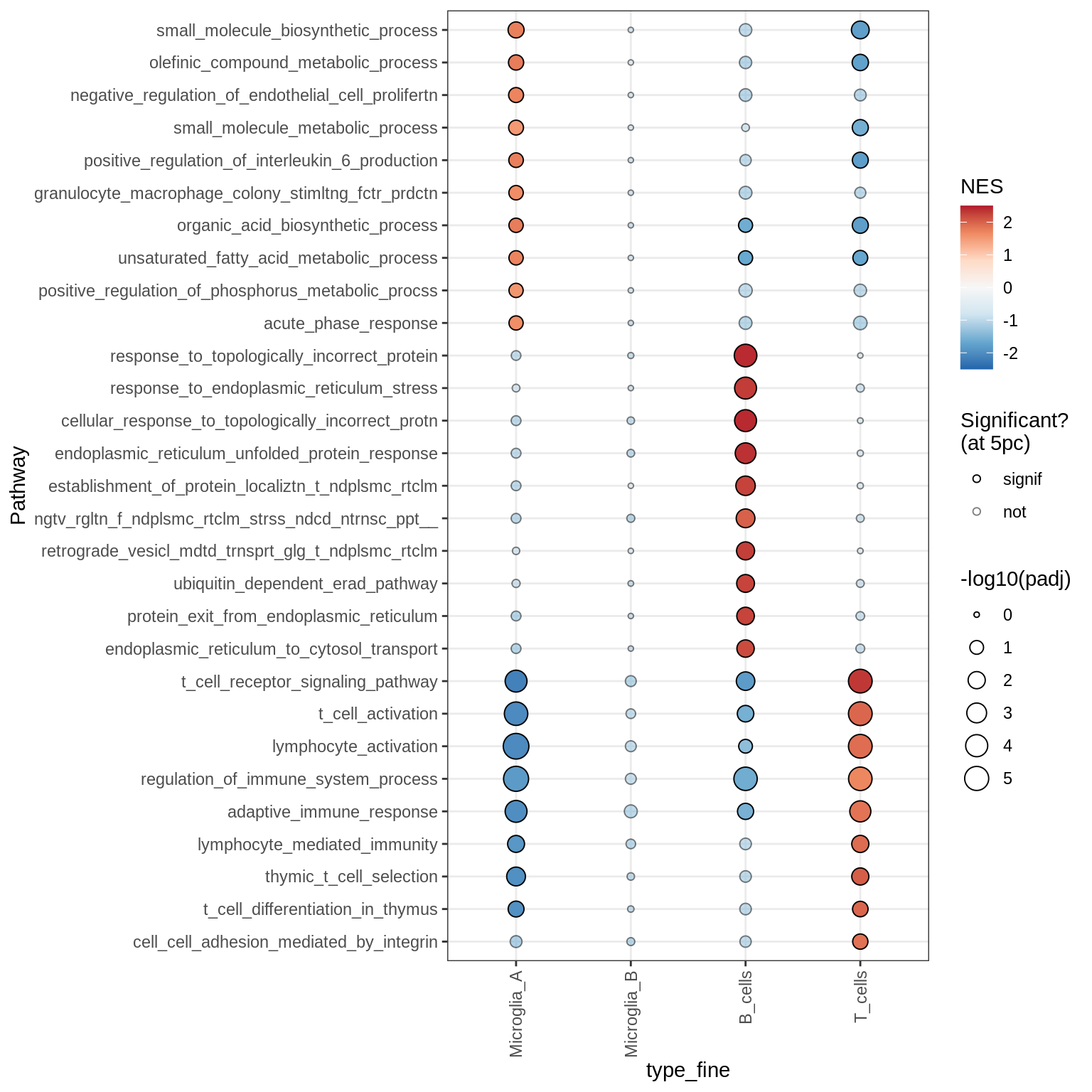
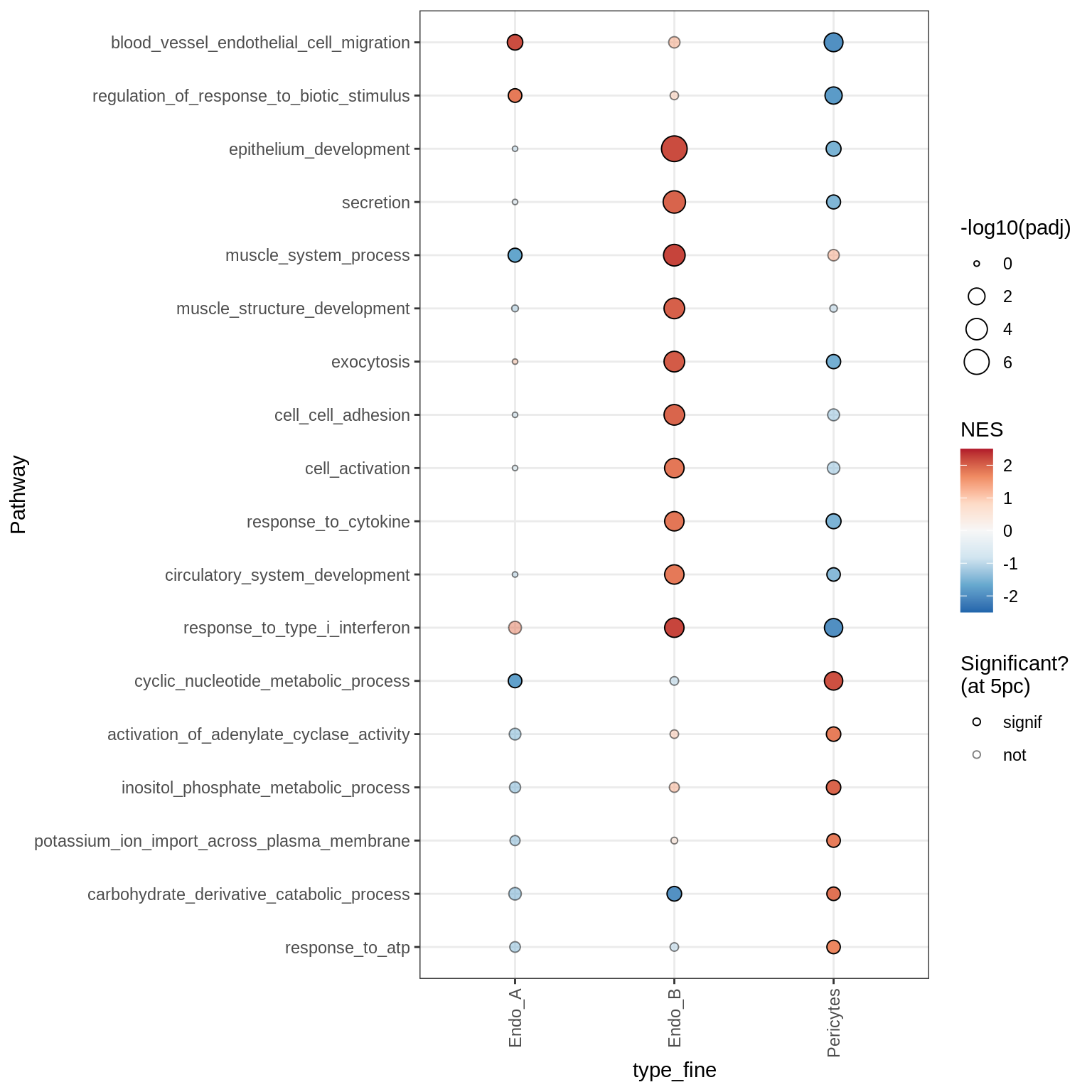
Dotplots of GSEA results (all samples, broad)
for (b in names(broad_list)) {
cat('### ', b, '\n')
print(plot_dotplot_gsea(fgsea_all, labels_dt, broad_list[[b]],
fgsea_cut = fgsea_cut))
cat('\n\n')
}OPCs / COPs
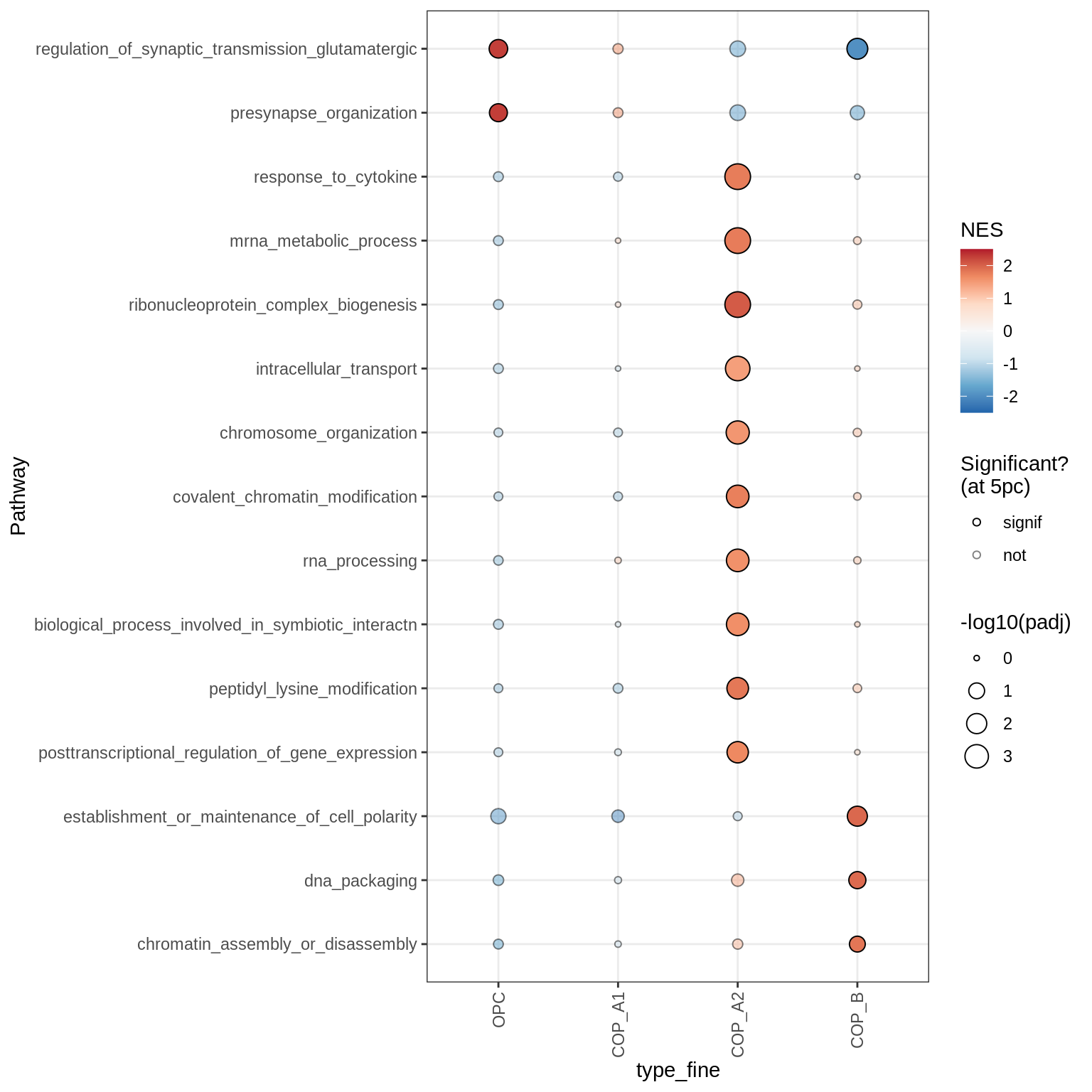
Oligodendrocytes
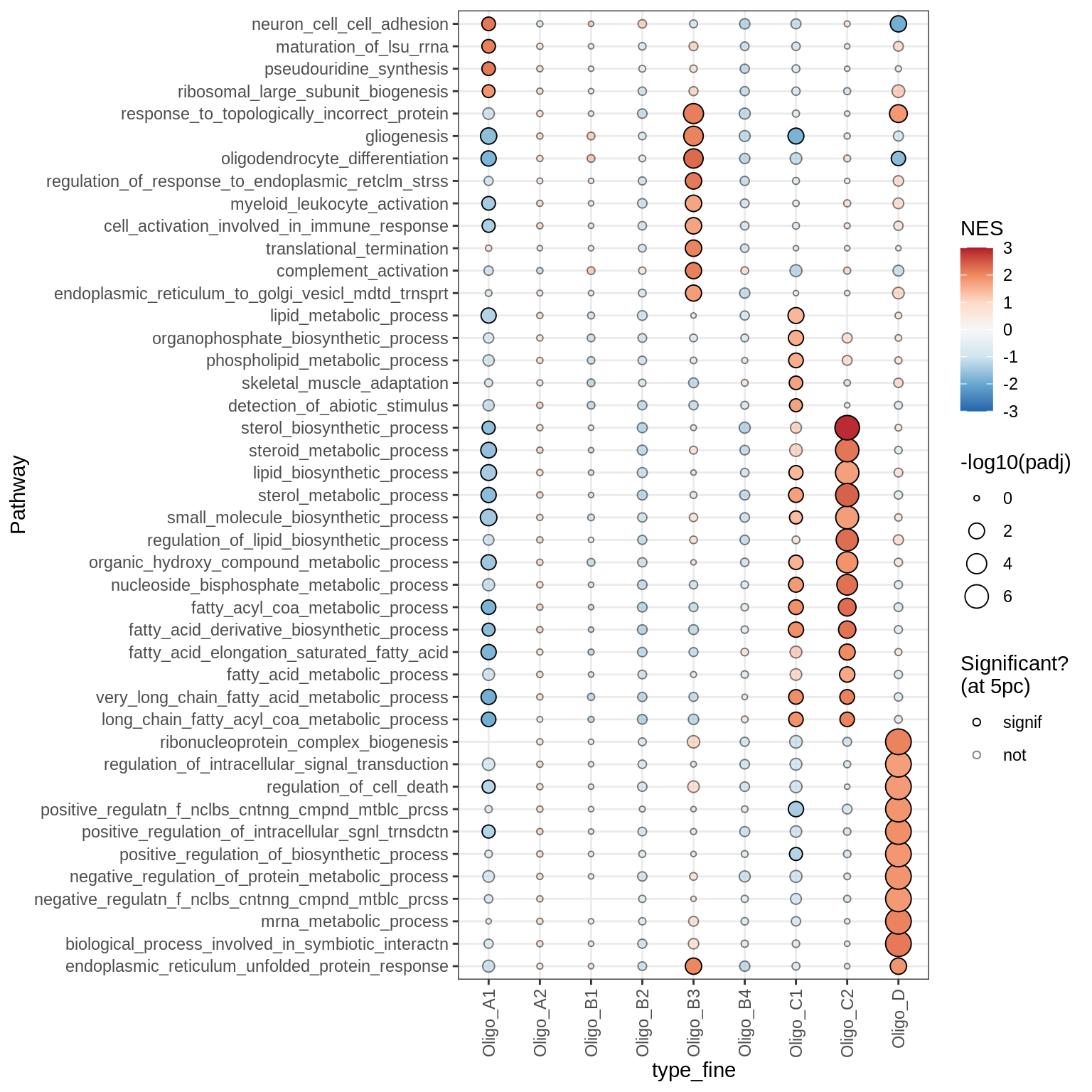
Astrocytes
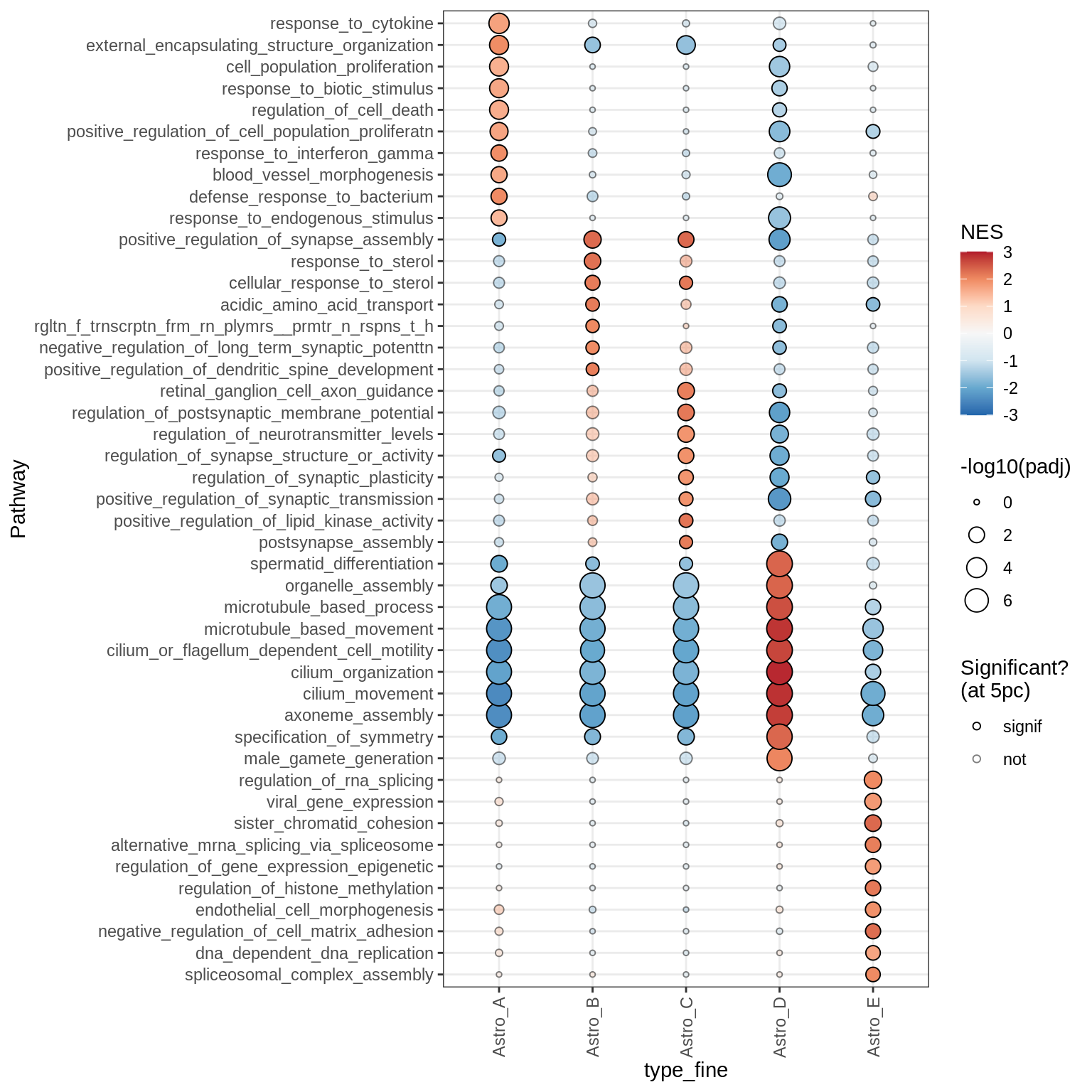
Microglia + Immune
Excitatory neurons
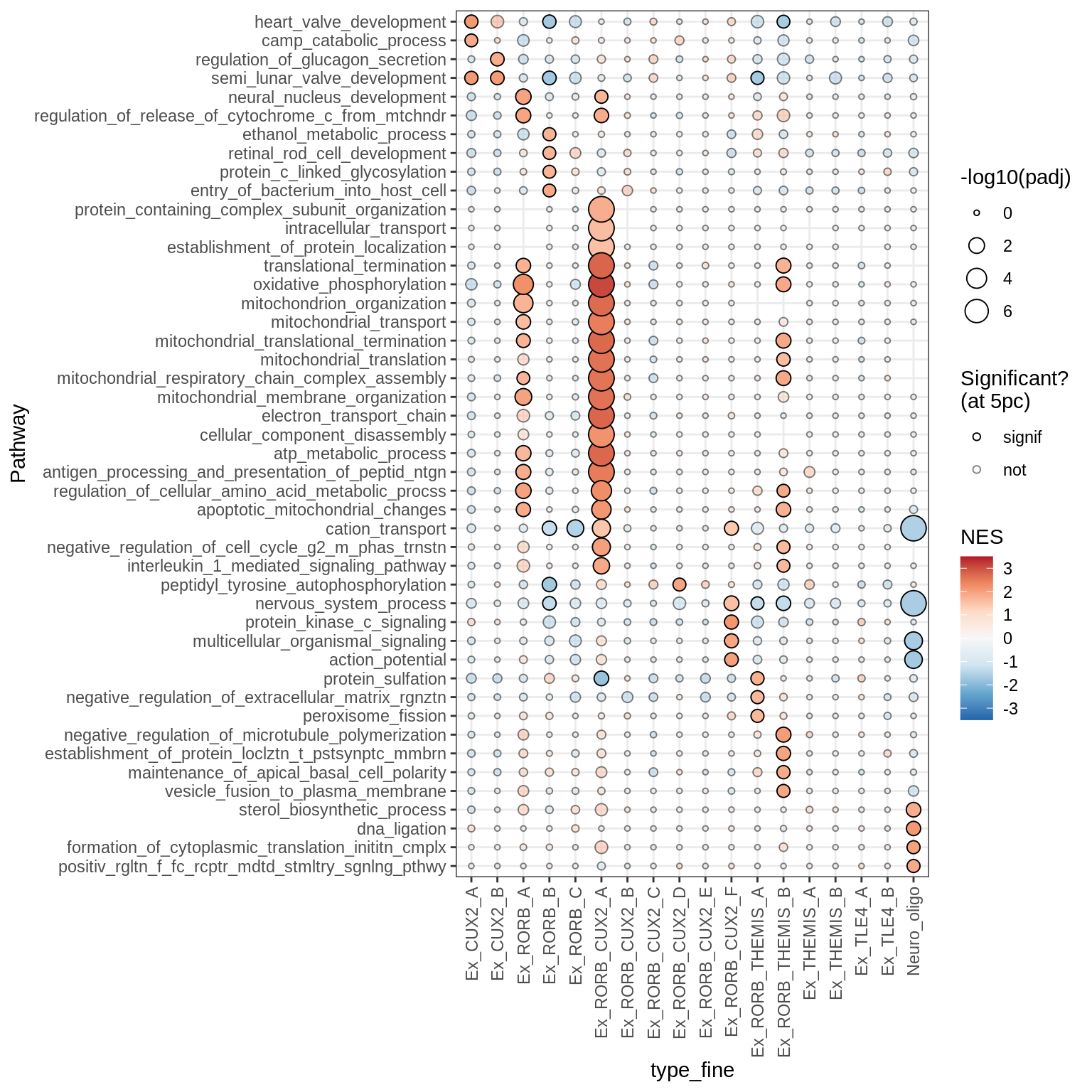
Inhibitory neurons
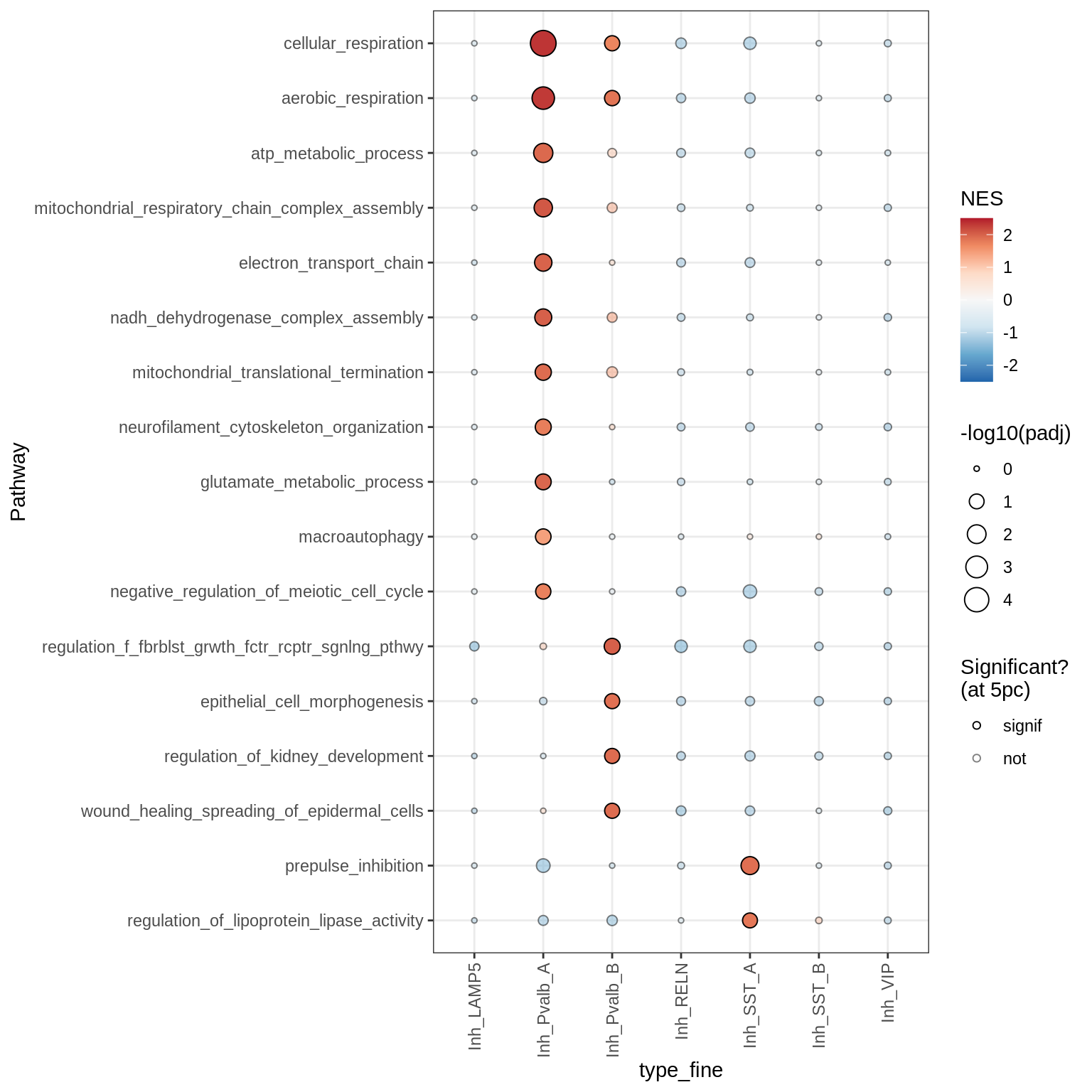
Endothelial + Pericytes
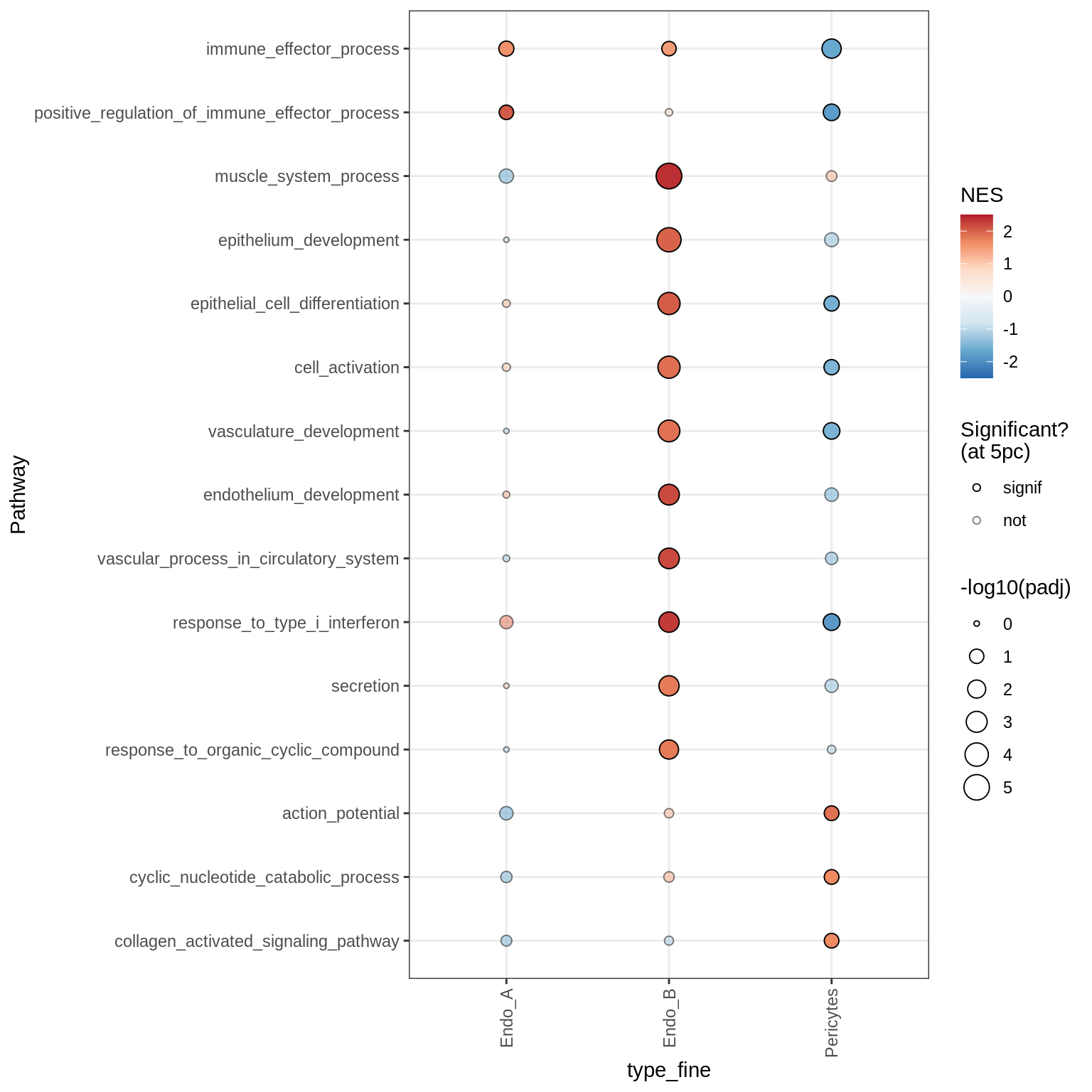
Dotplots of GSEA results (healthy samples, broad)
for (b in names(broad_list)) {
cat('### ', b, '\n')
print(plot_dotplot_gsea(fgsea_ctrl, labels_dt, broad_list[[b]],
fgsea_cut = fgsea_cut))
cat('\n\n')
}OPCs / COPs
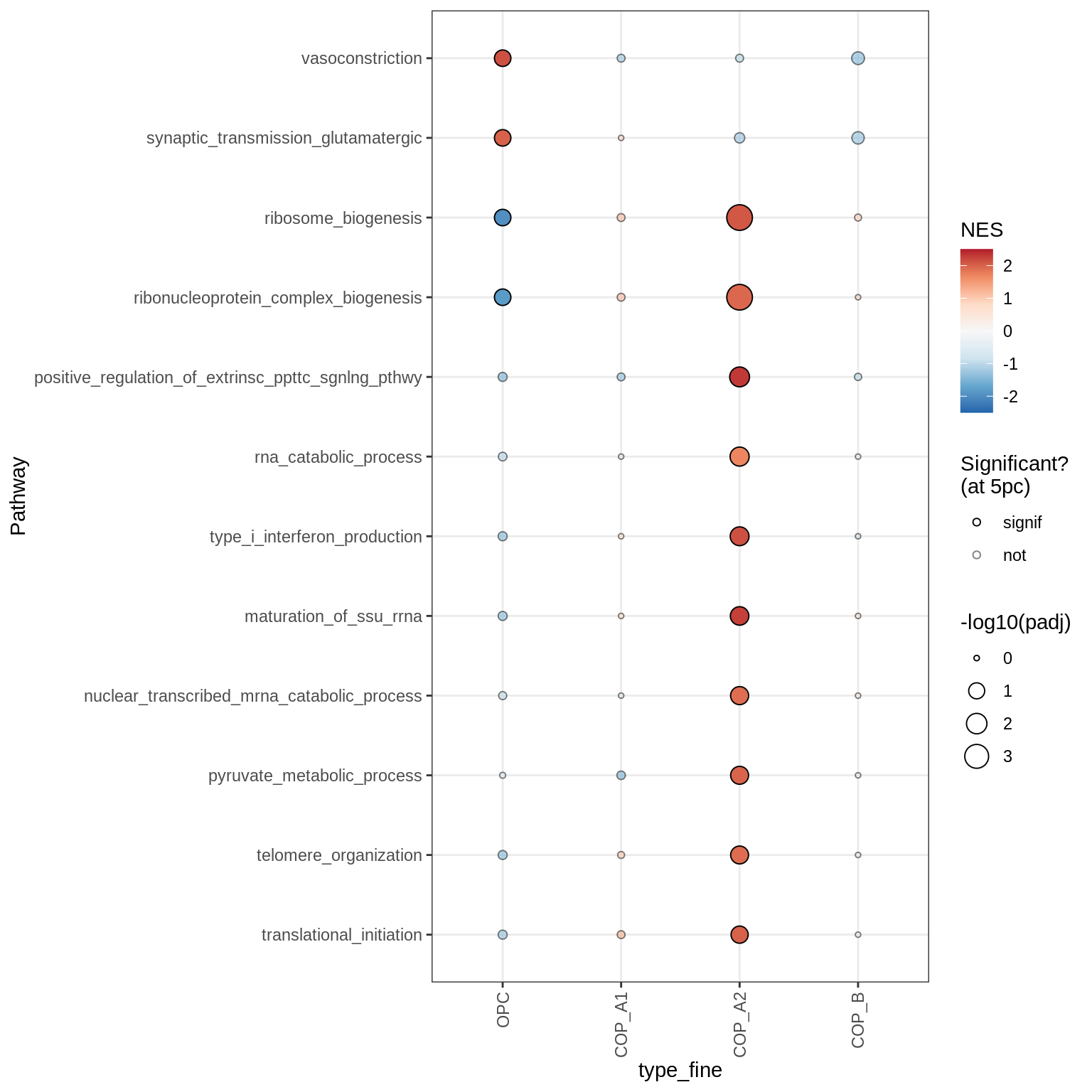
Oligodendrocytes
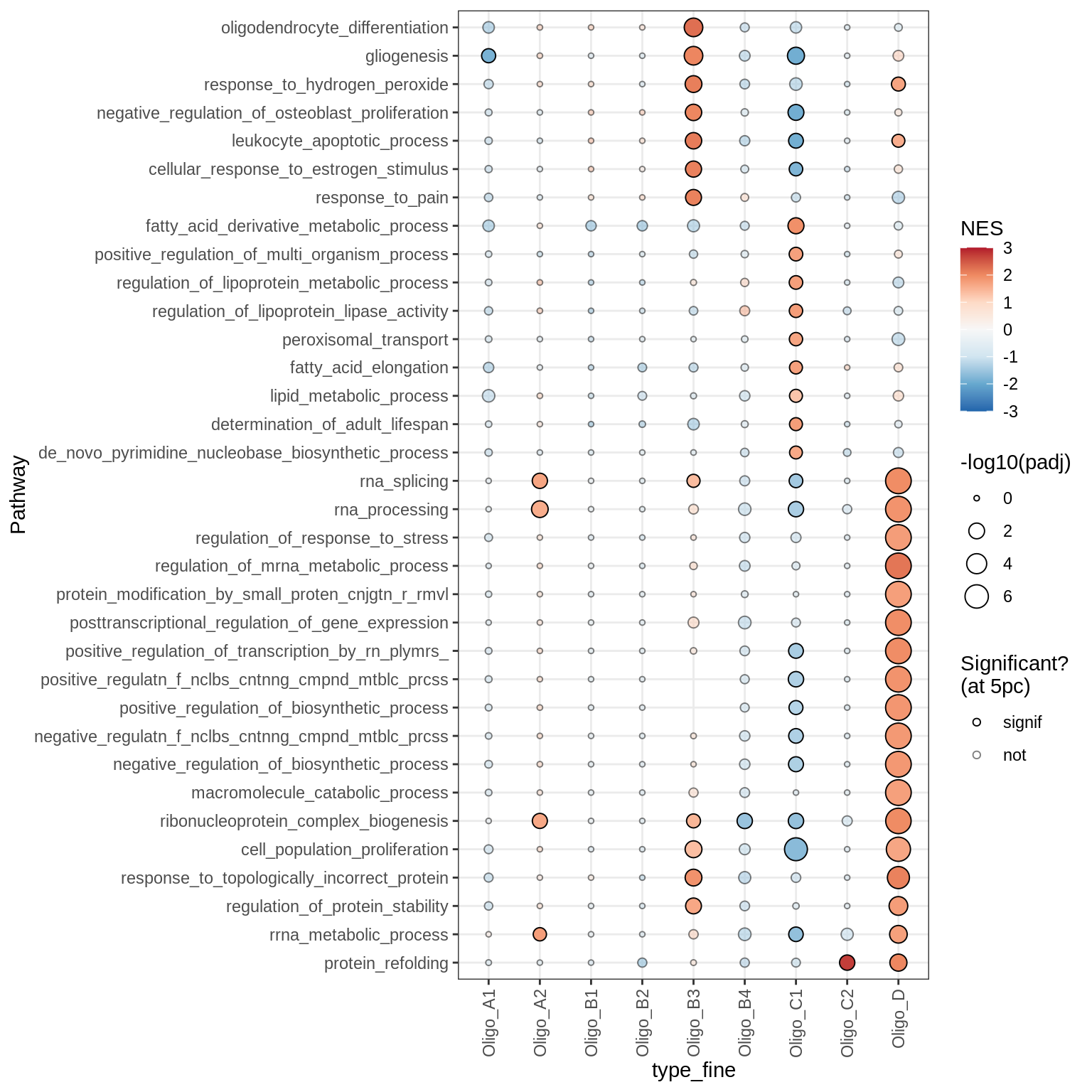
Astrocytes
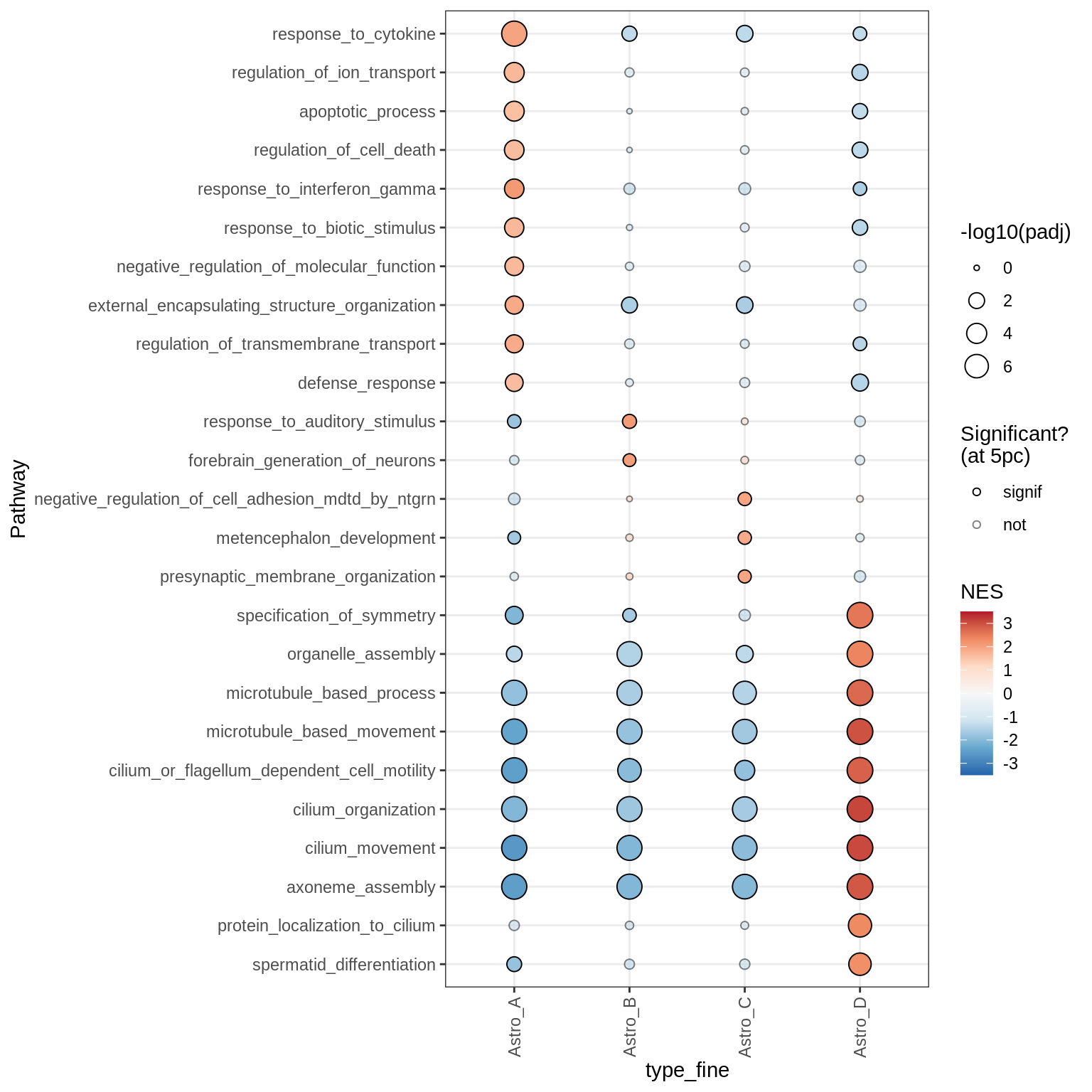
Microglia + Immune
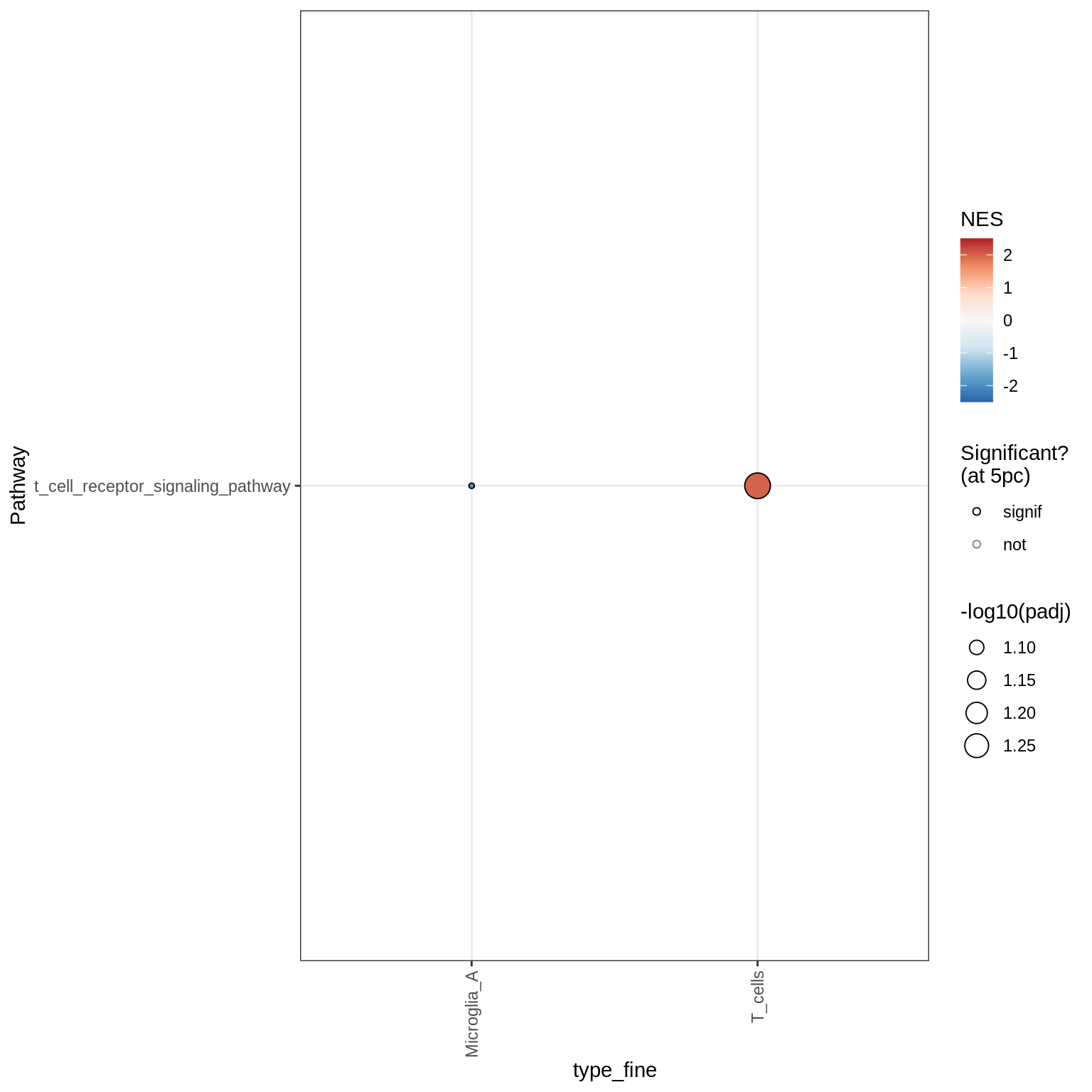
Excitatory neurons

Inhibitory neurons
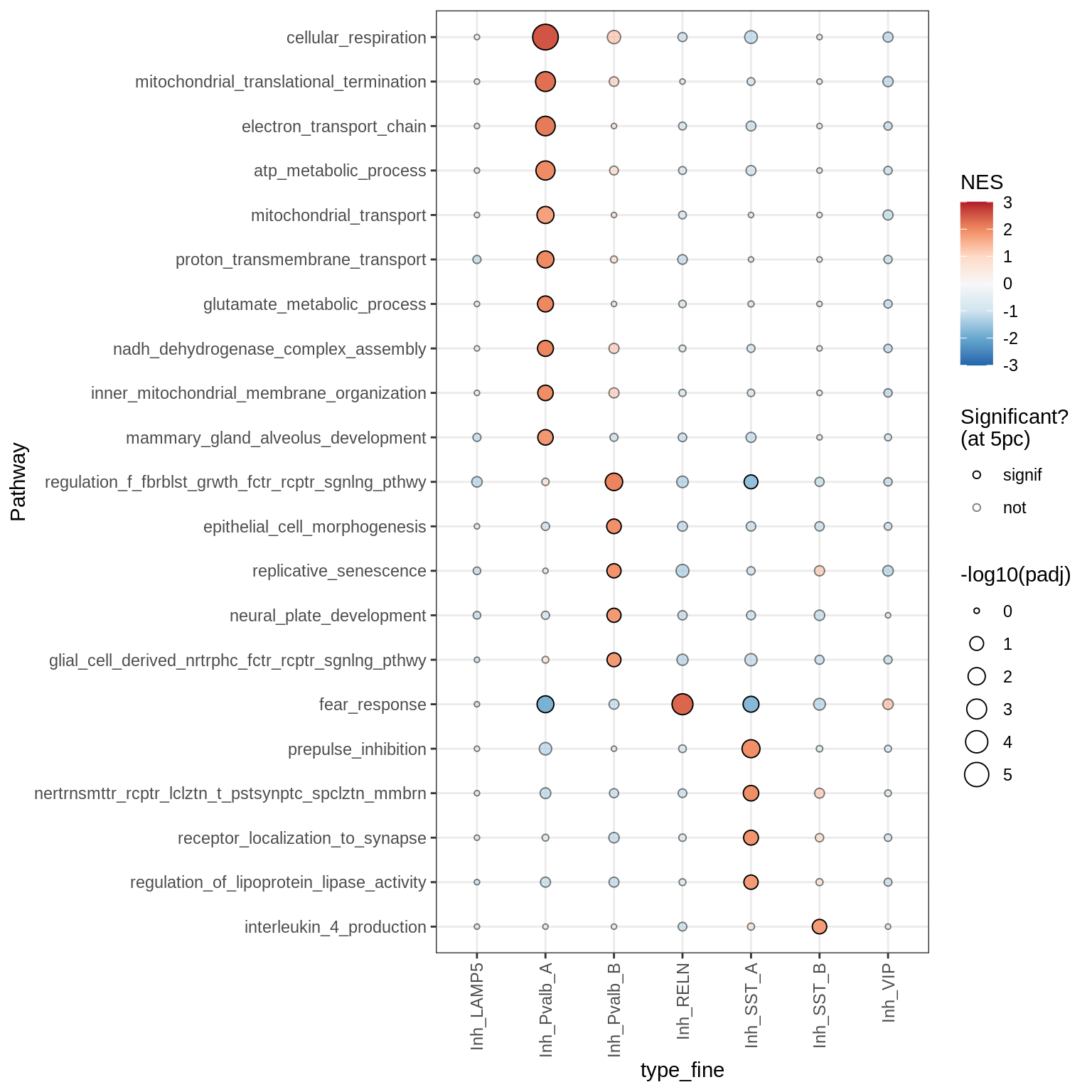
Endothelial + Pericytes
Dotplot of microglia / macrophage genes
suppressWarnings(print(plot_dotplot(pb_fine, props_fine, micro_gs, labels_dt,
row_split = 'micro_class')))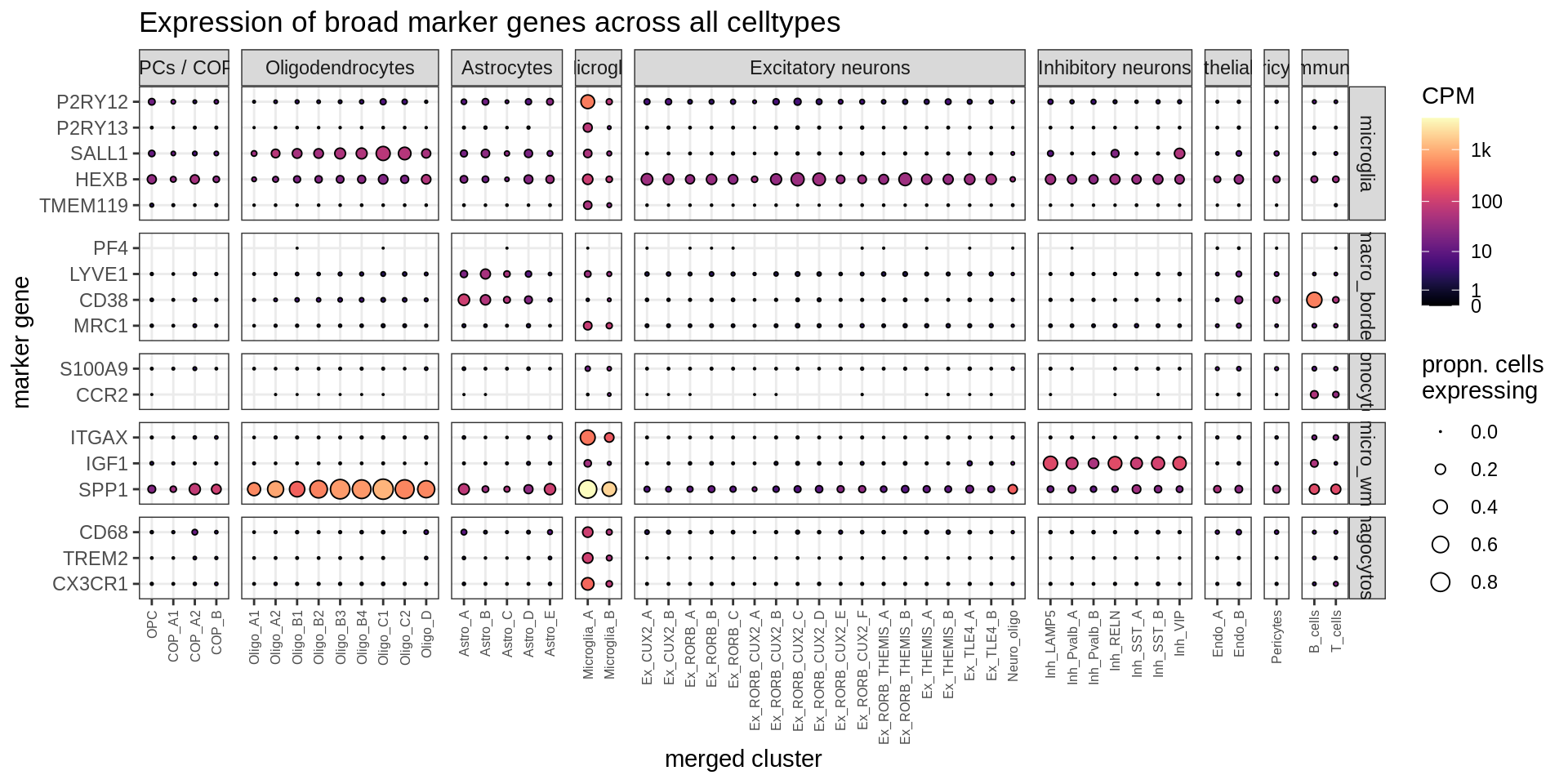
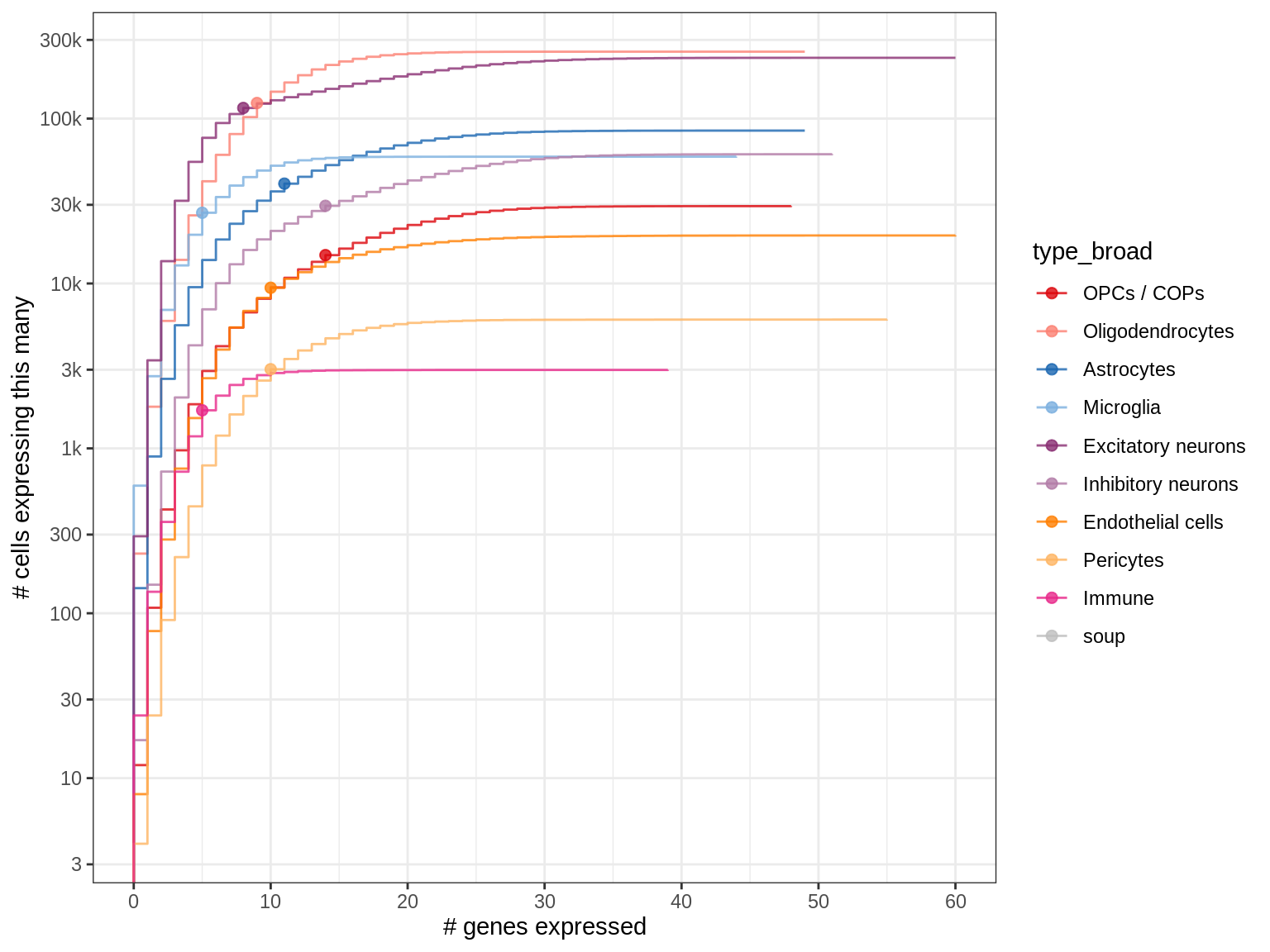
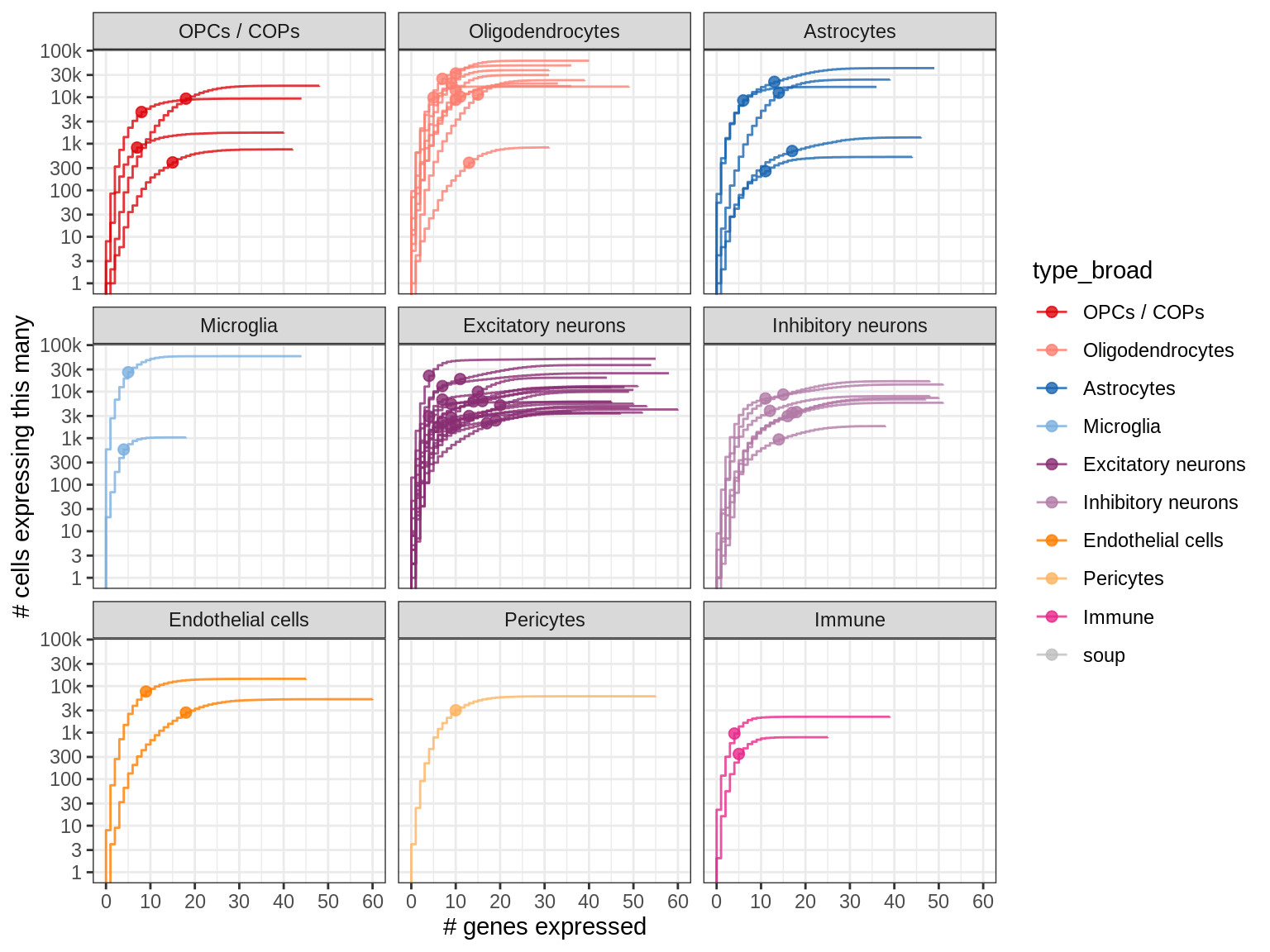
How frequently are panel markers expressed in different celltypes?
The dots show the median expressing cell, i.e. 50% of the cells of a given type express this many genes or less. So for example, 50% of the Immune cells in our snRNAseq dataset express at most 4 of the panel genes.
for (what_level in c('type_broad', 'type_fine')) {
cat('### ', what_level, '\n')
suppressWarnings(print(plot_binary_distributions(n_exp_dt, what_level, dot_cut = 0.5)))
cat('\n\n')
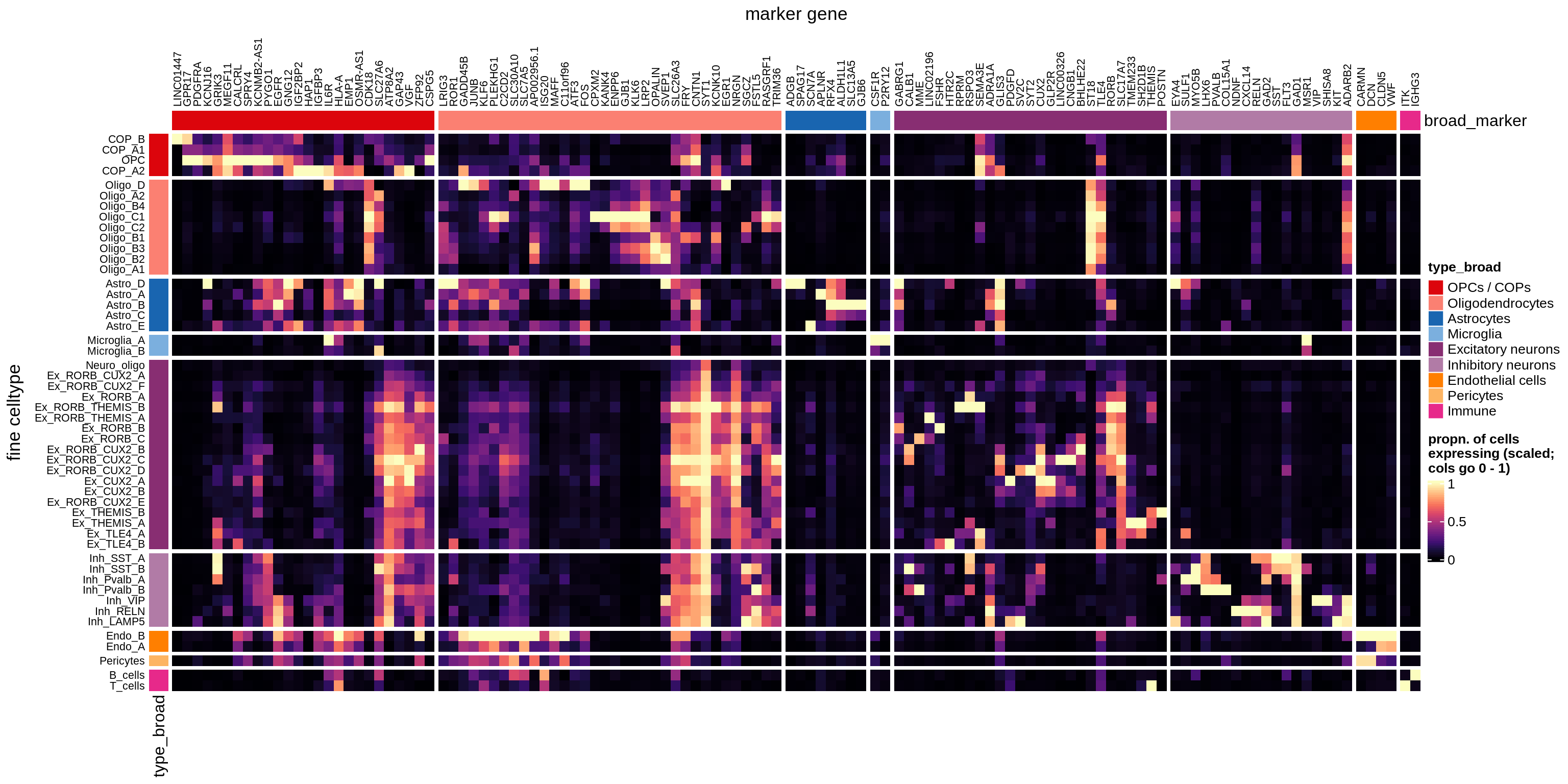
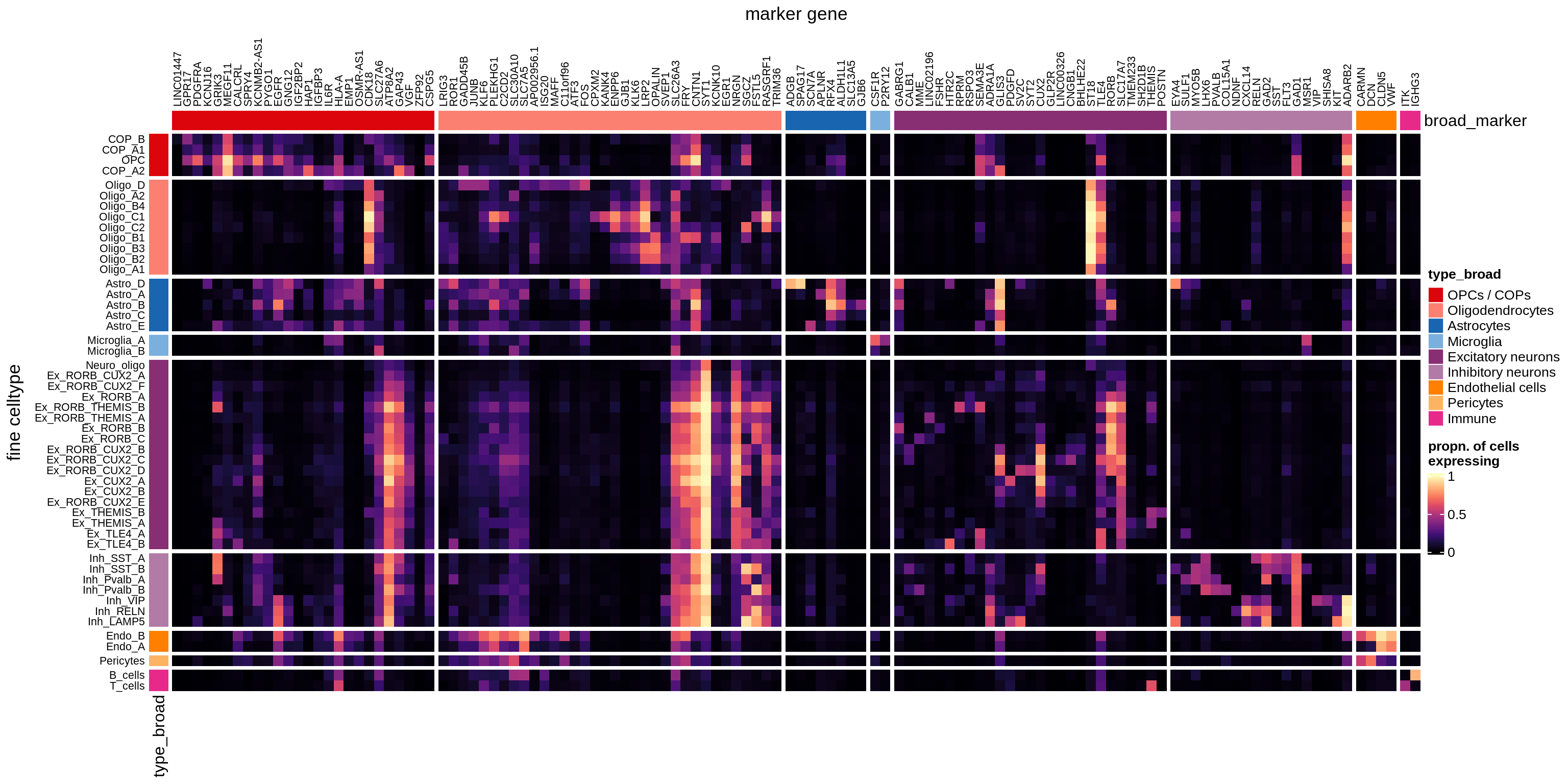
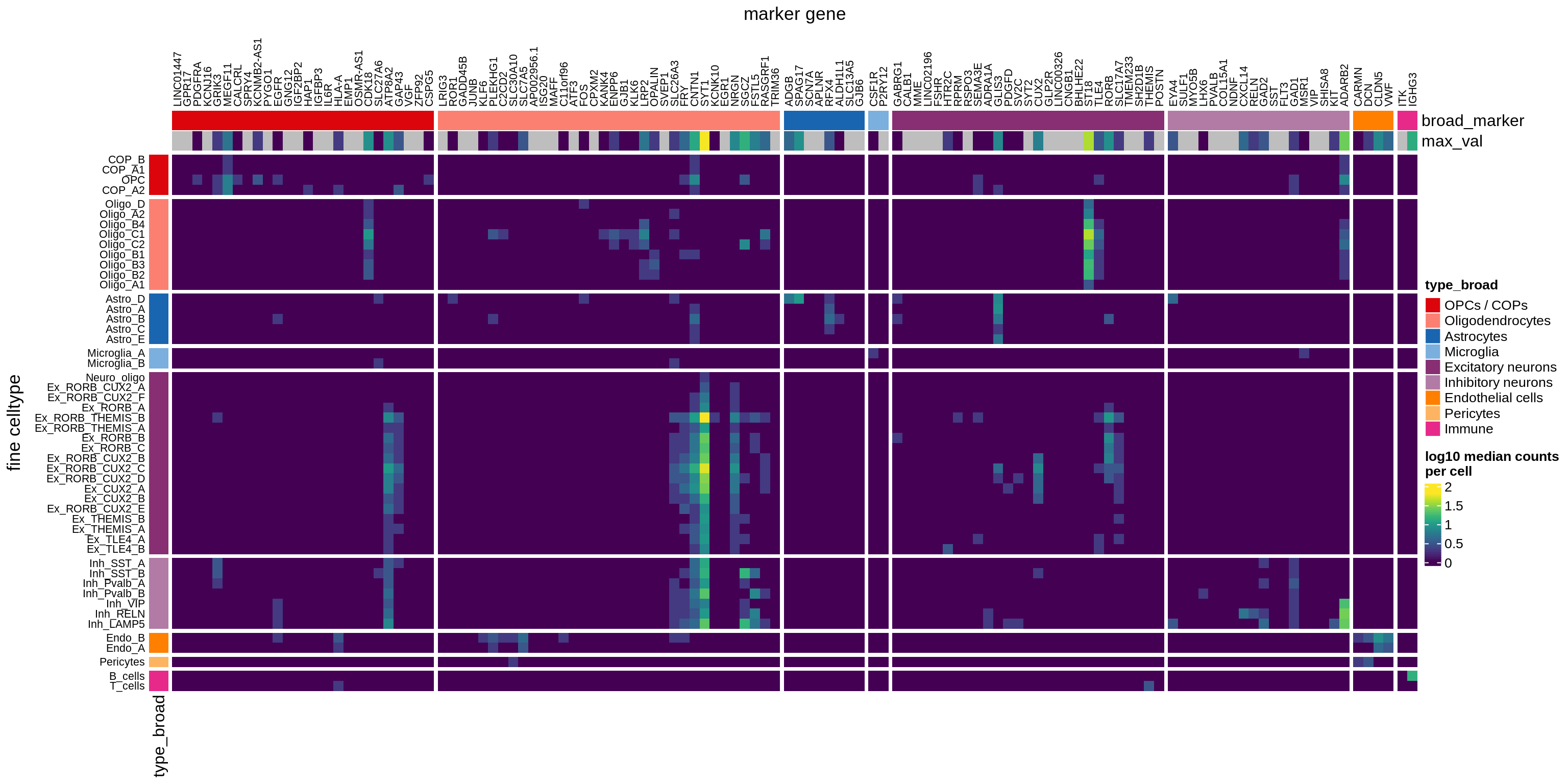
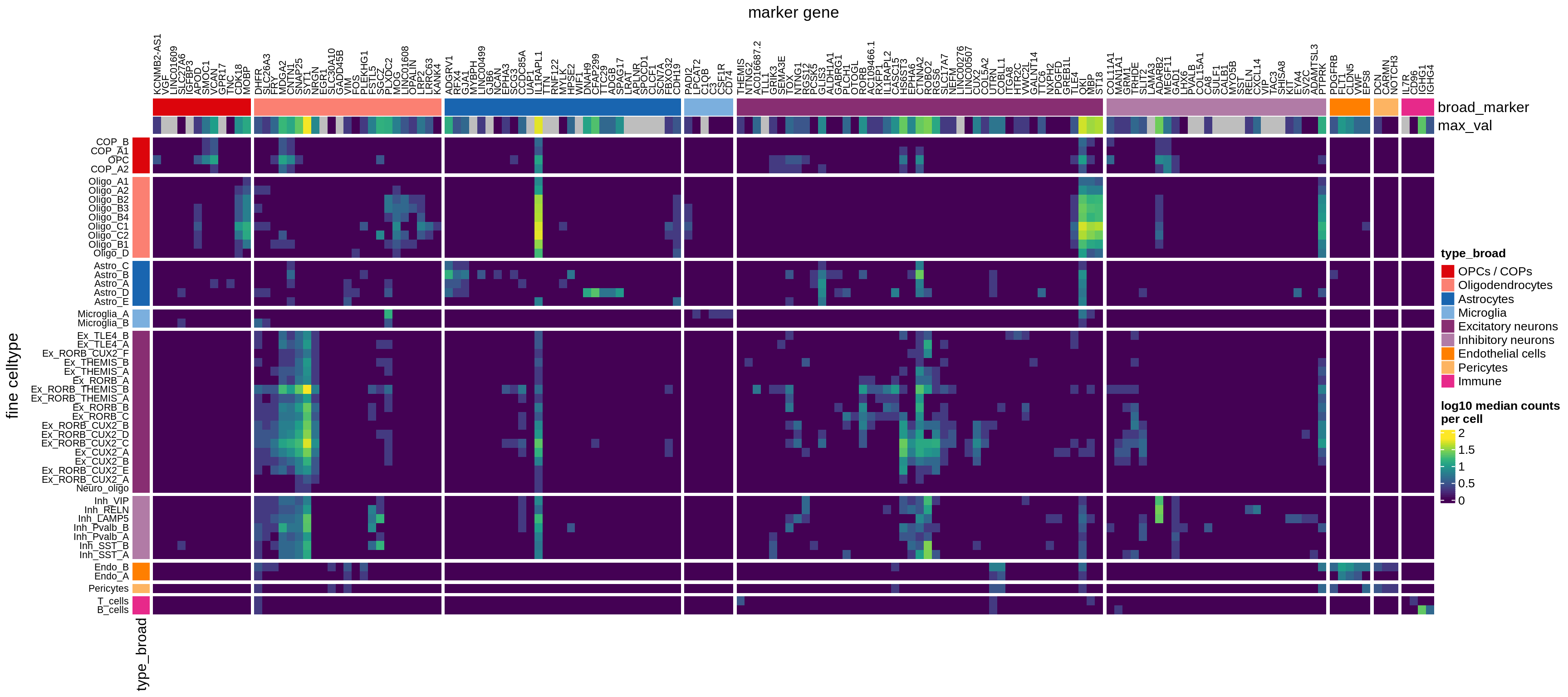
}Heatmaps of panel marker expression - Cartana panel
what_list = c(
'Propn. of cells expressing (scaled)',
'Propn. of cells expressing',
'Median counts per cell'
) %>% setNames(c('mean_scaled', 'mean_exp', 'median_exp'))
for (what in names(what_list)) {
cat('### ', what_list[[what]], '\n')
if (what == 'median_exp') {
suppressWarnings(print(plot_marker_heatmap(medians_dt, labels_dt,
seriate_obj, what)))
} else {
suppressWarnings(print(plot_marker_heatmap(prop_exp_dt, labels_dt,
seriate_obj, what)))
}
cat('\n\n')
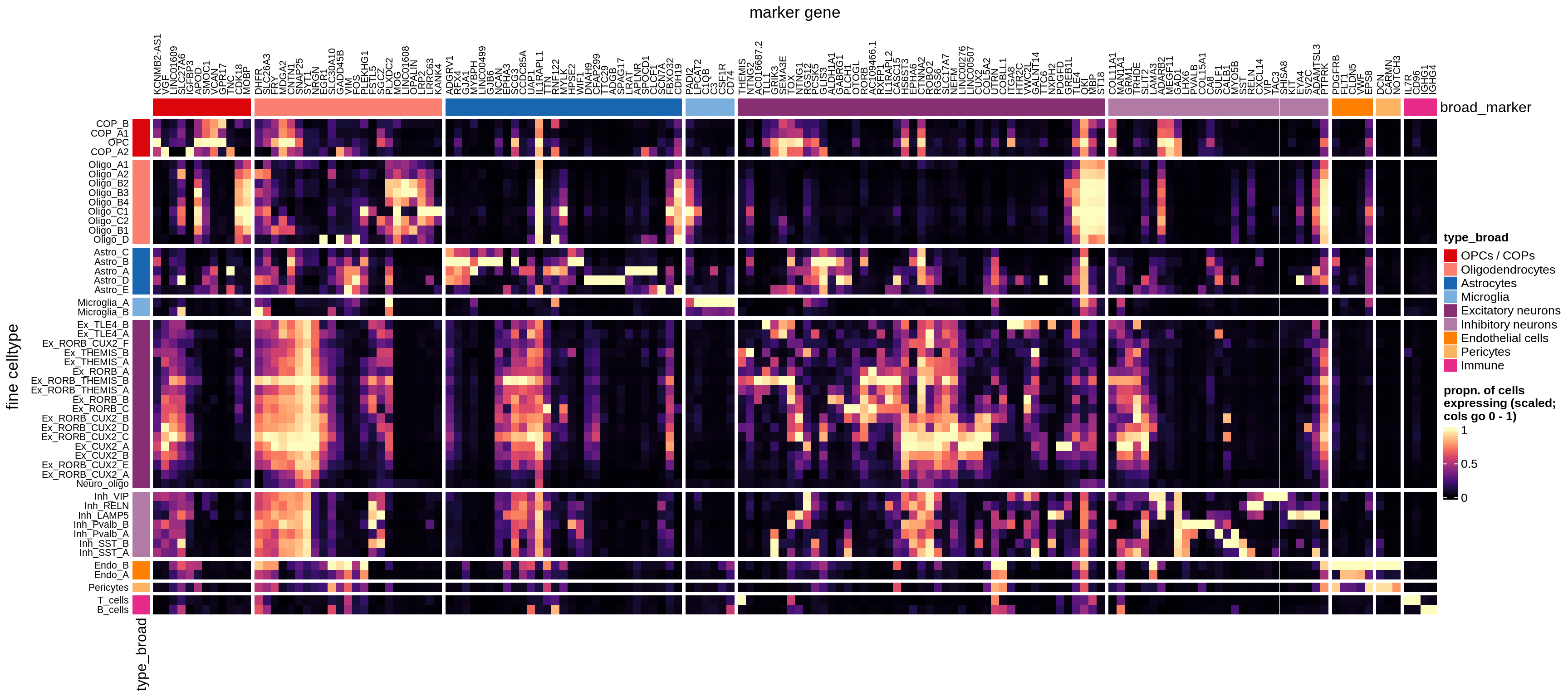
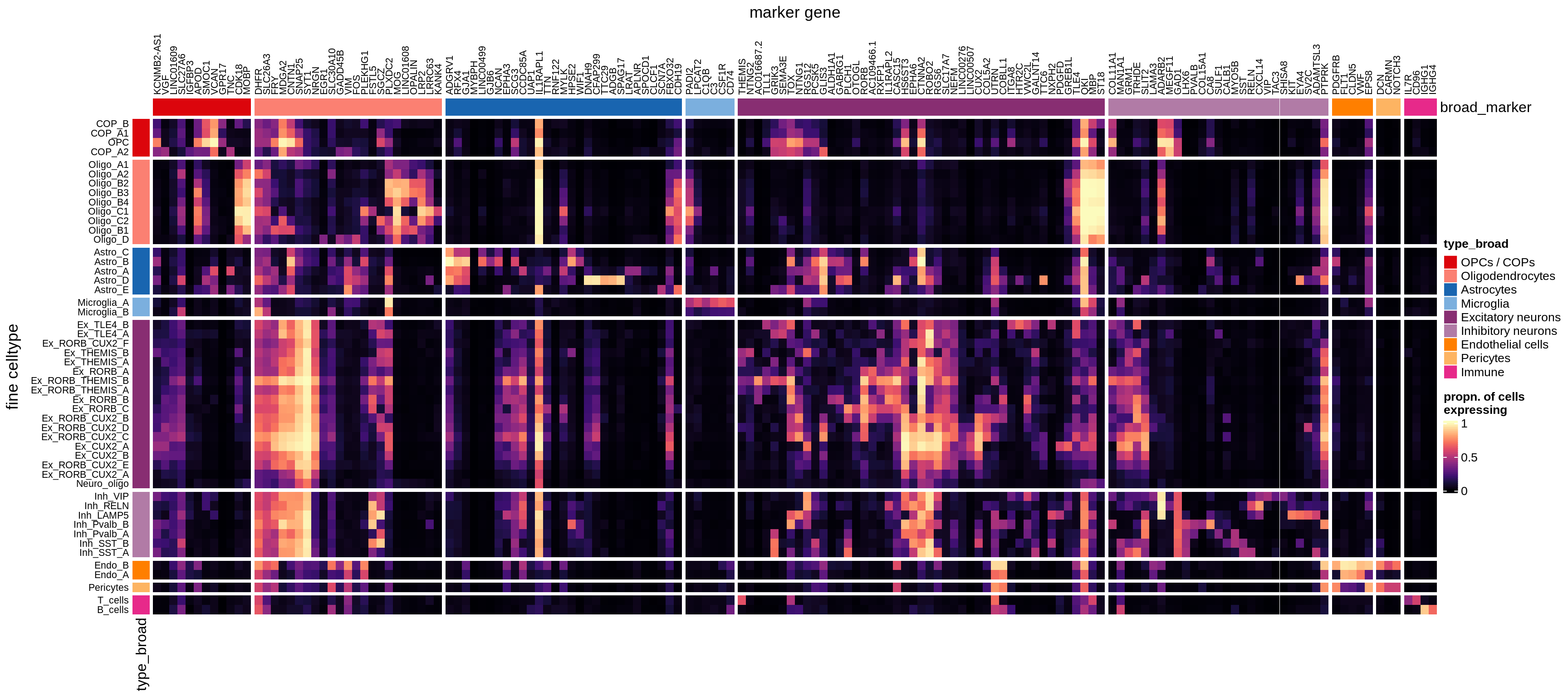
}Heatmaps of panel marker expression - Roche panel
for (what in names(what_list)) {
cat('### ', what_list[[what]], '\n')
if (what == 'median_exp') {
suppressWarnings(print(plot_marker_heatmap(medians_dt_roche, labels_dt,
seriate_roche, what)))
} else {
suppressWarnings(print(plot_marker_heatmap(prop_exp_dt_roche, labels_dt,
seriate_roche, what)))
}
cat('\n\n')
}Classifier using panel markers
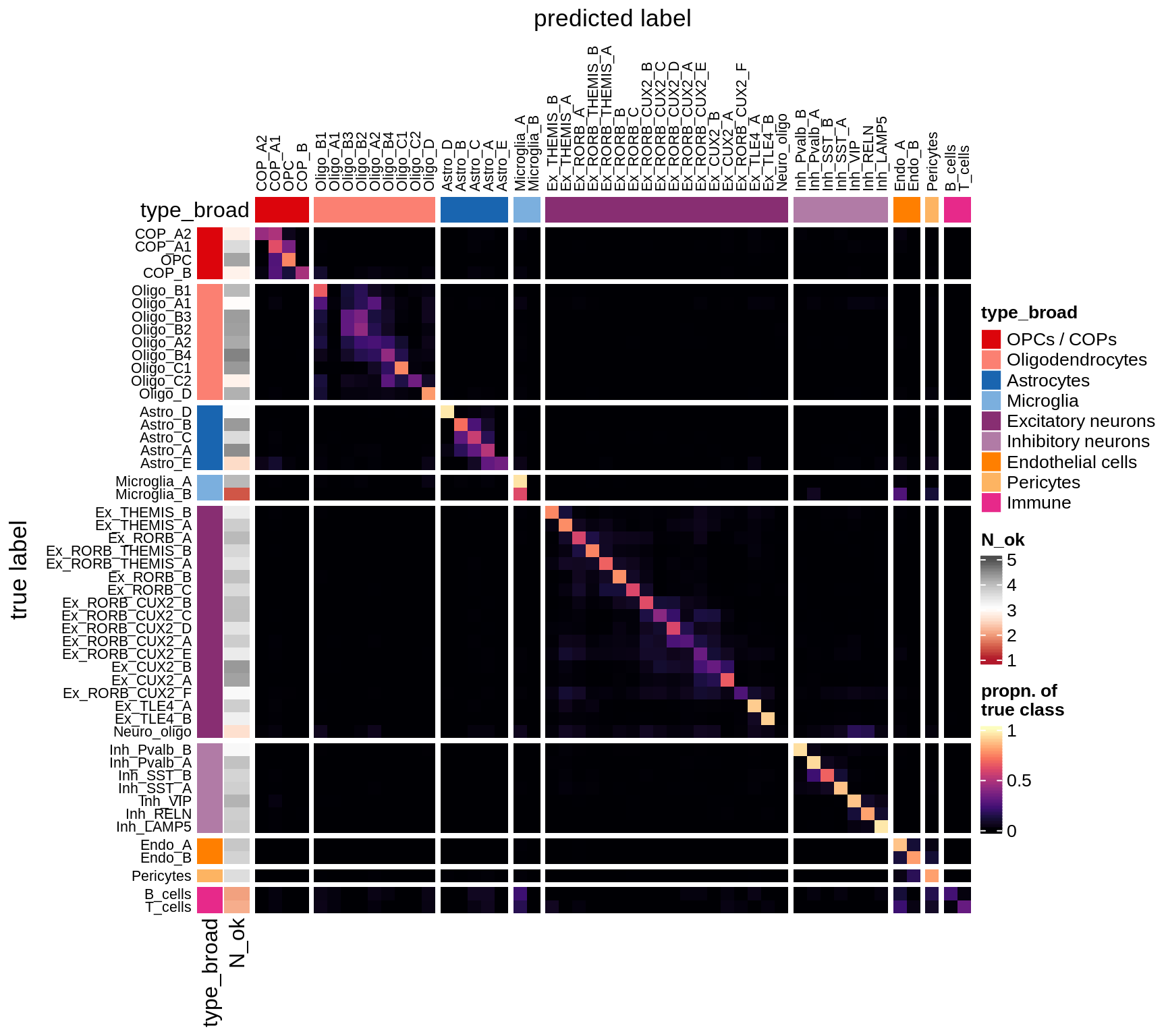
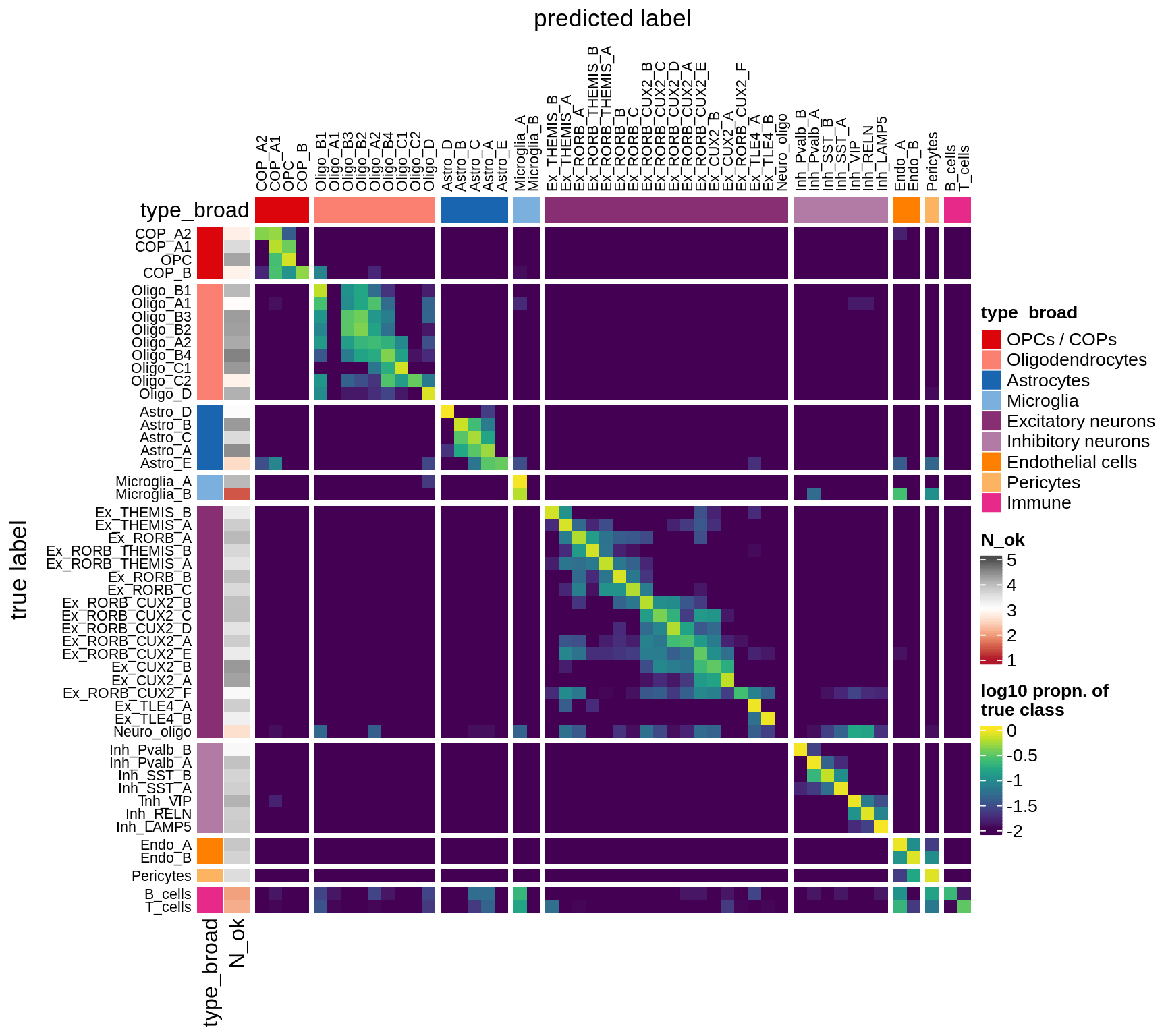
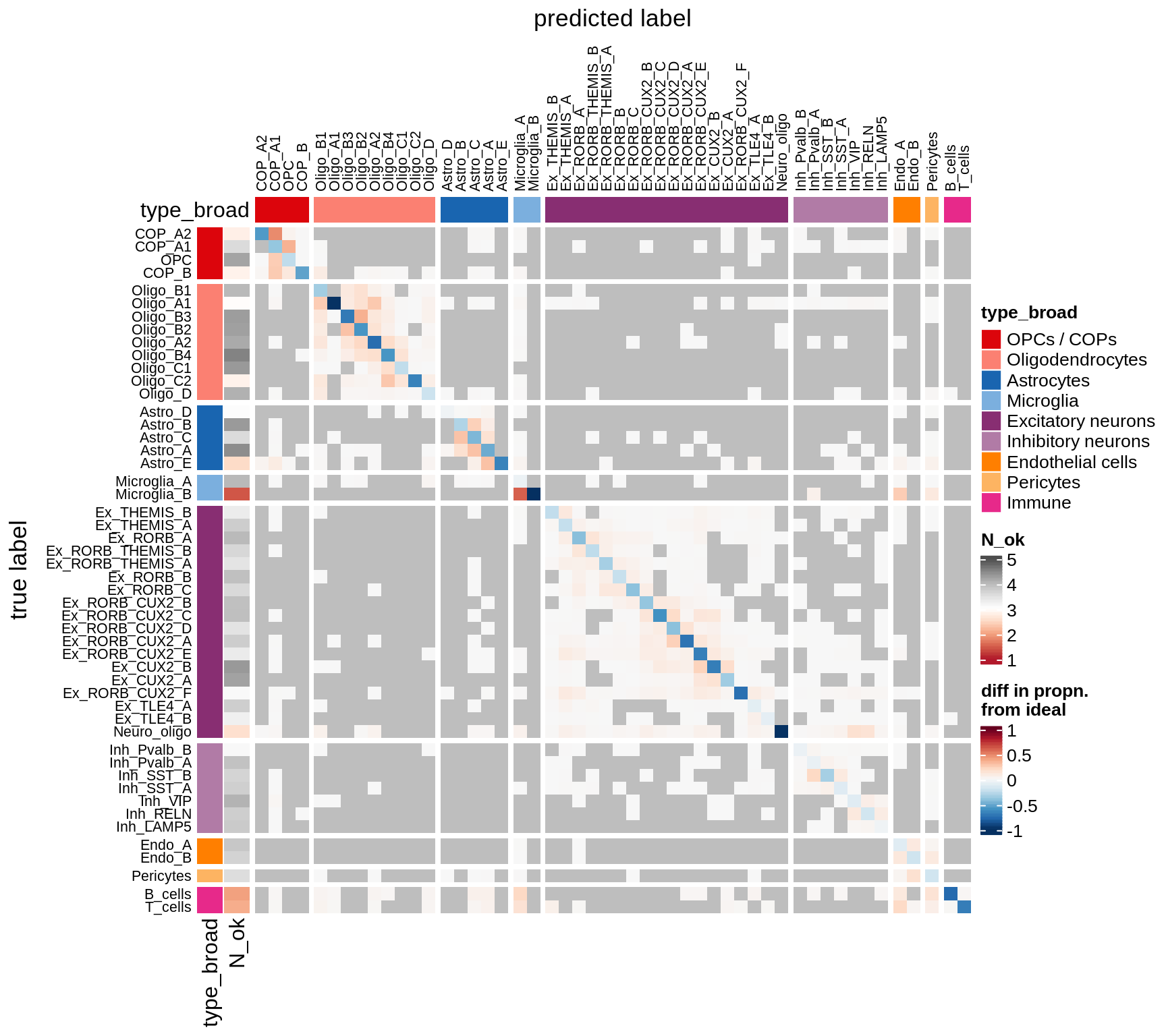
I took 1k cells per celltype, and trained a classifier using binary gene expression (i.e. we just see a 0 or a 1 for each panel gene in each cell). Then I tested this on another 1k cells per celltype. This gives an idea of how easy it is to identify these celltypes with our panel.
The three heatmaps show:
- prob Predictions for each celltype, in proportions, so each row sums to 1. In a perfect world, this would be diagonal, indicating that each celltype was predicted only to be that celltype. The N_ok column on the left shows how many cells had at least 10 panel genes expressed, and that we could use in the classifier.
- logp Same as 1., but with log values of the proportions.
- diff This how far away from the perfect world we are. You see red where this prediction is made too much, e.g. immune cells being predicted as microglia. You see blue where predictions are missing, so for all Immune cells the correct celltype is not predicted sufficiently.
for (what in c('prob', 'logp', 'diff')) {
cat('### ', what, '\n')
suppressWarnings(draw(plot_classifier_heatmap(conf_dt, labels_dt, what)))
cat('\n\n')
}Conos using panel markers - clusters
draw(plot_conos_check(conos_dt, conos_chk))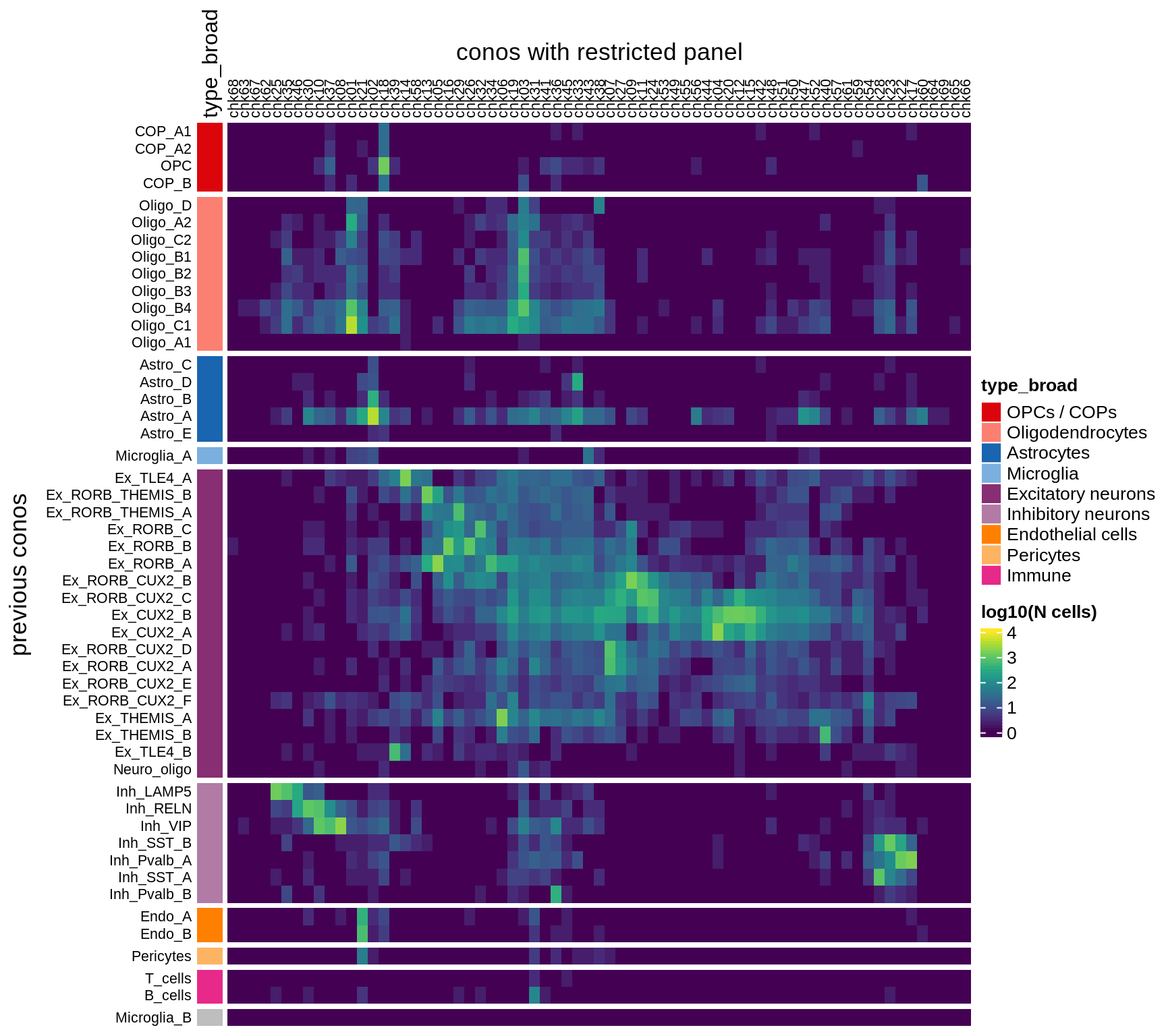
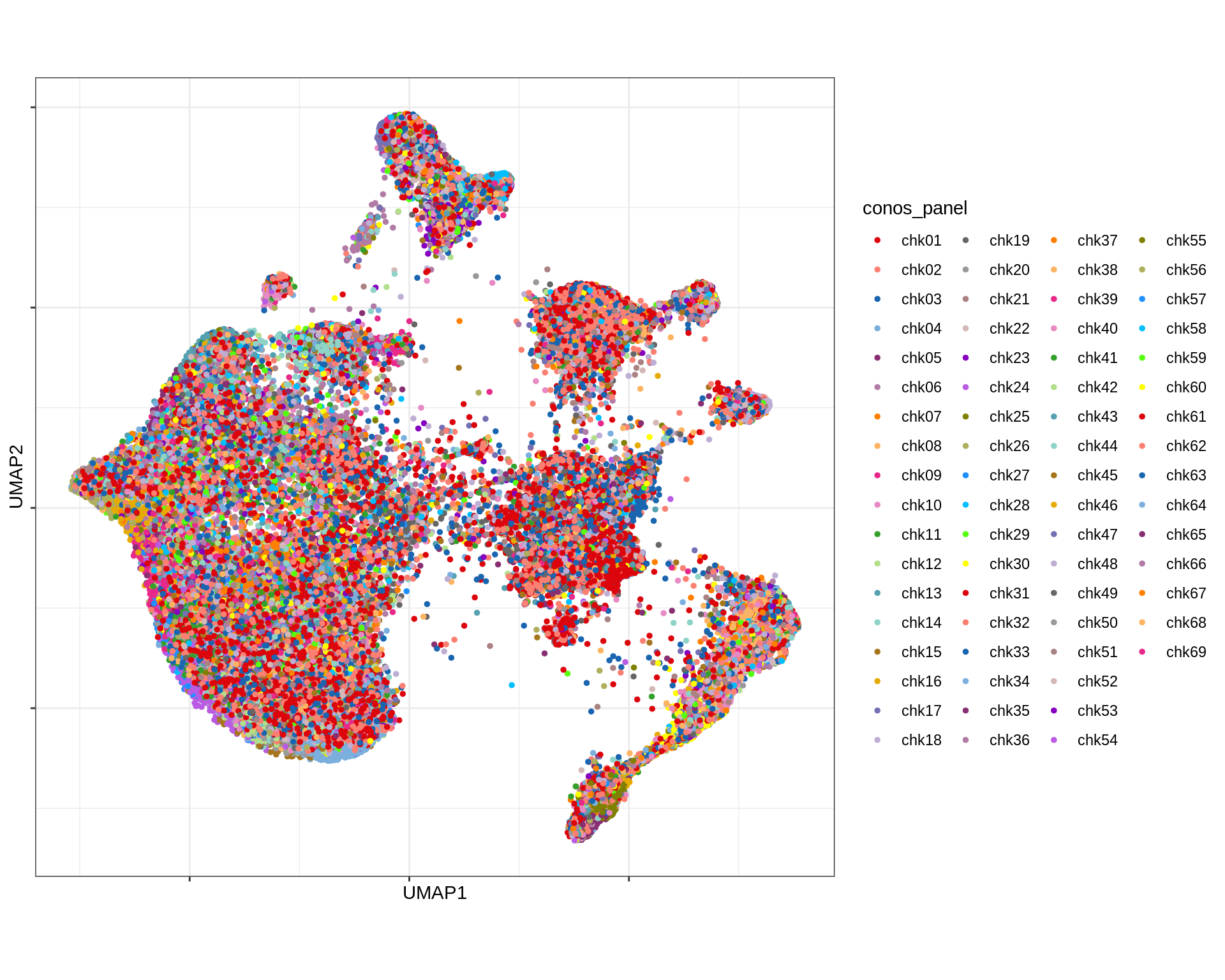
Conos using panel markers - UMAP
for (what in c('type_broad', 'type_fine', 'conos_panel')) {
cat('### ', what, '\n')
print(plot_conos_umap(conos_chk, conos_dt, what))
cat('\n\n')
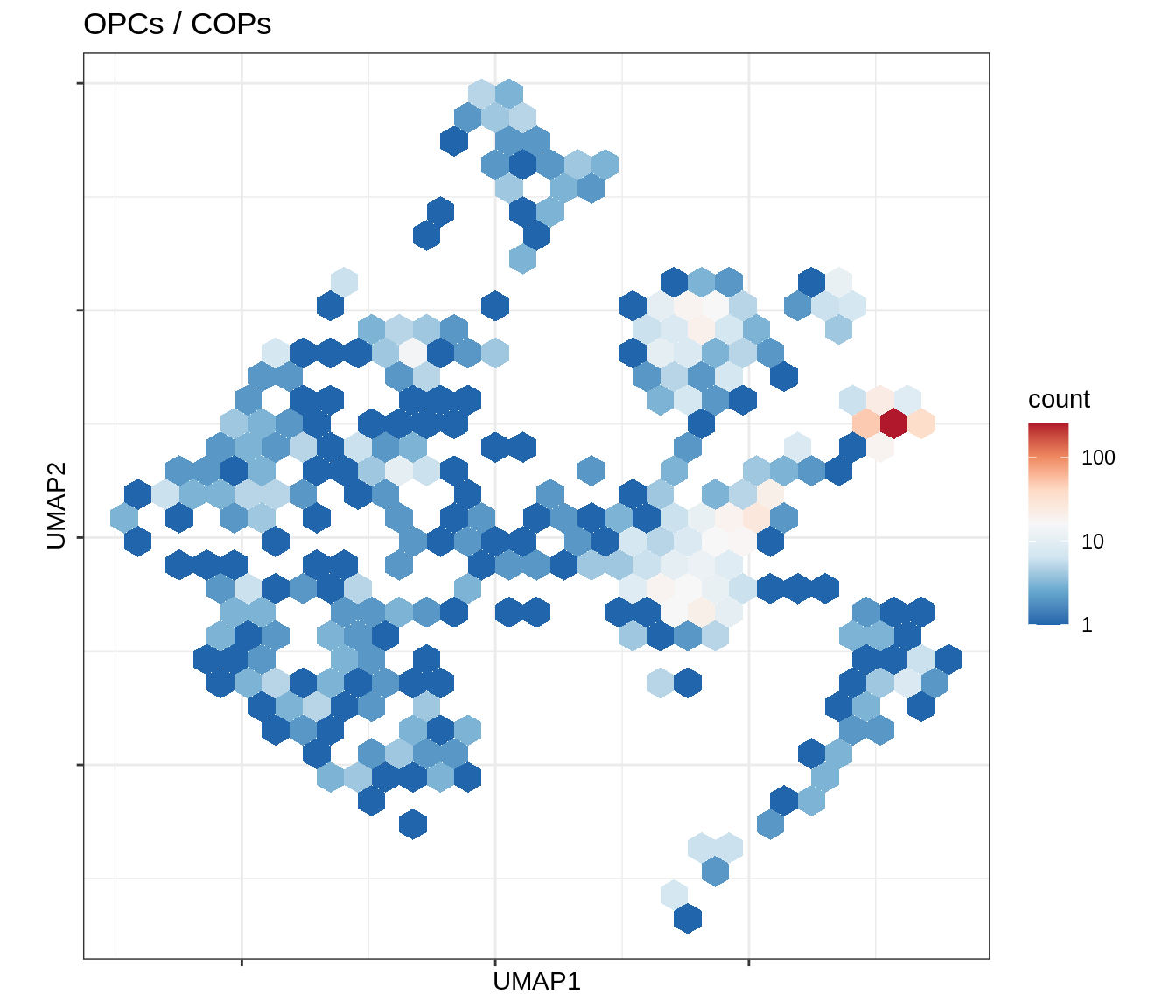
}Conos using panel markers - UMAP density
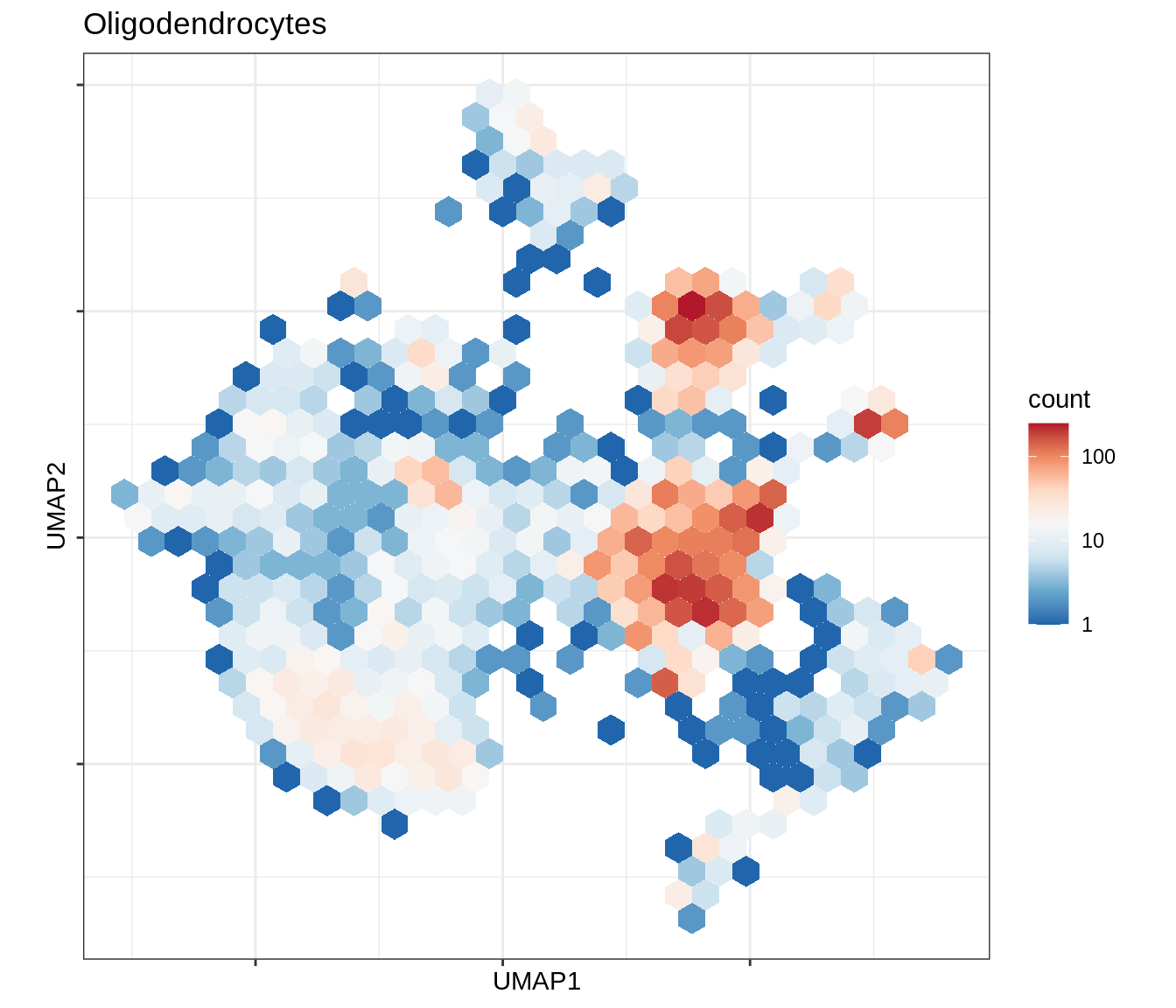
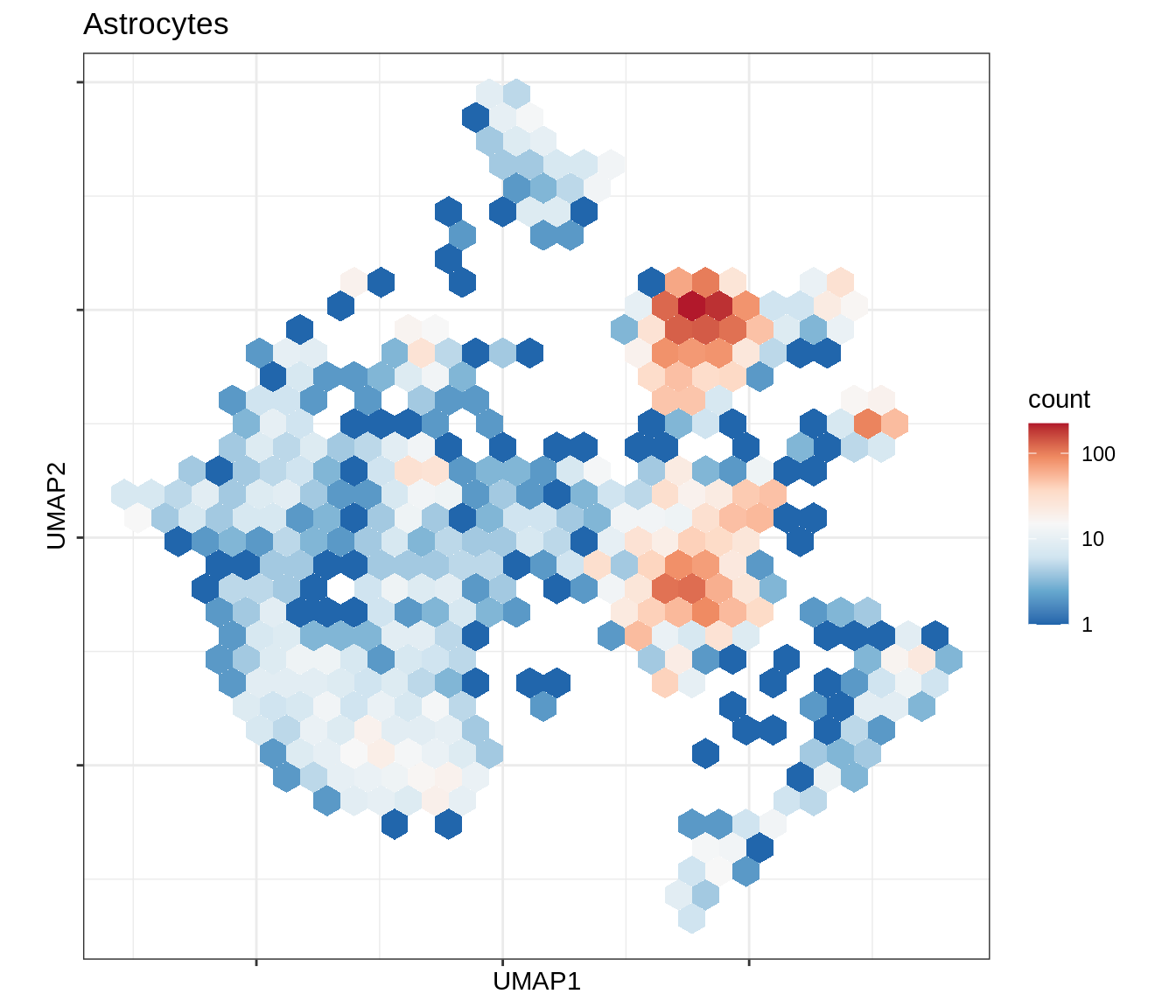
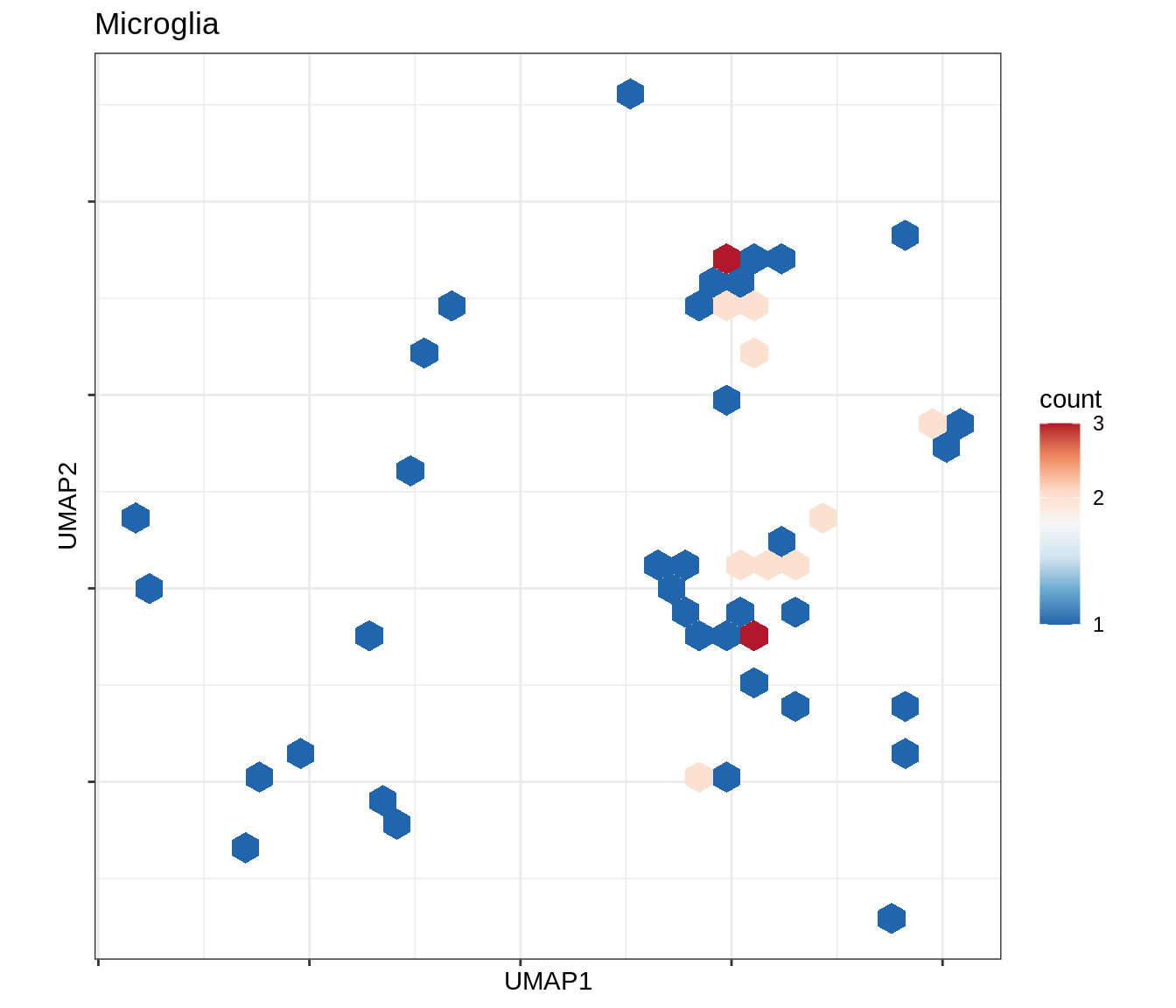
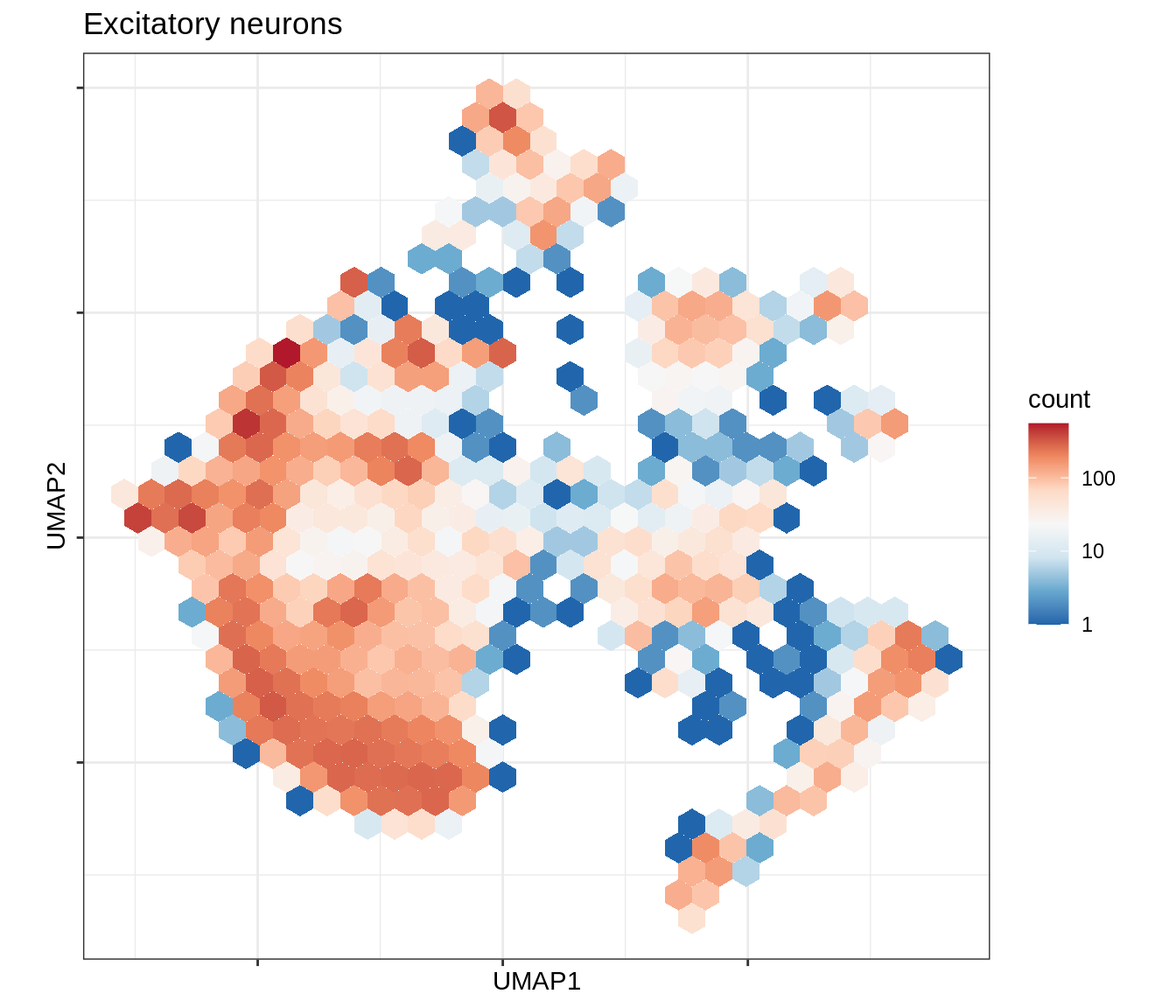
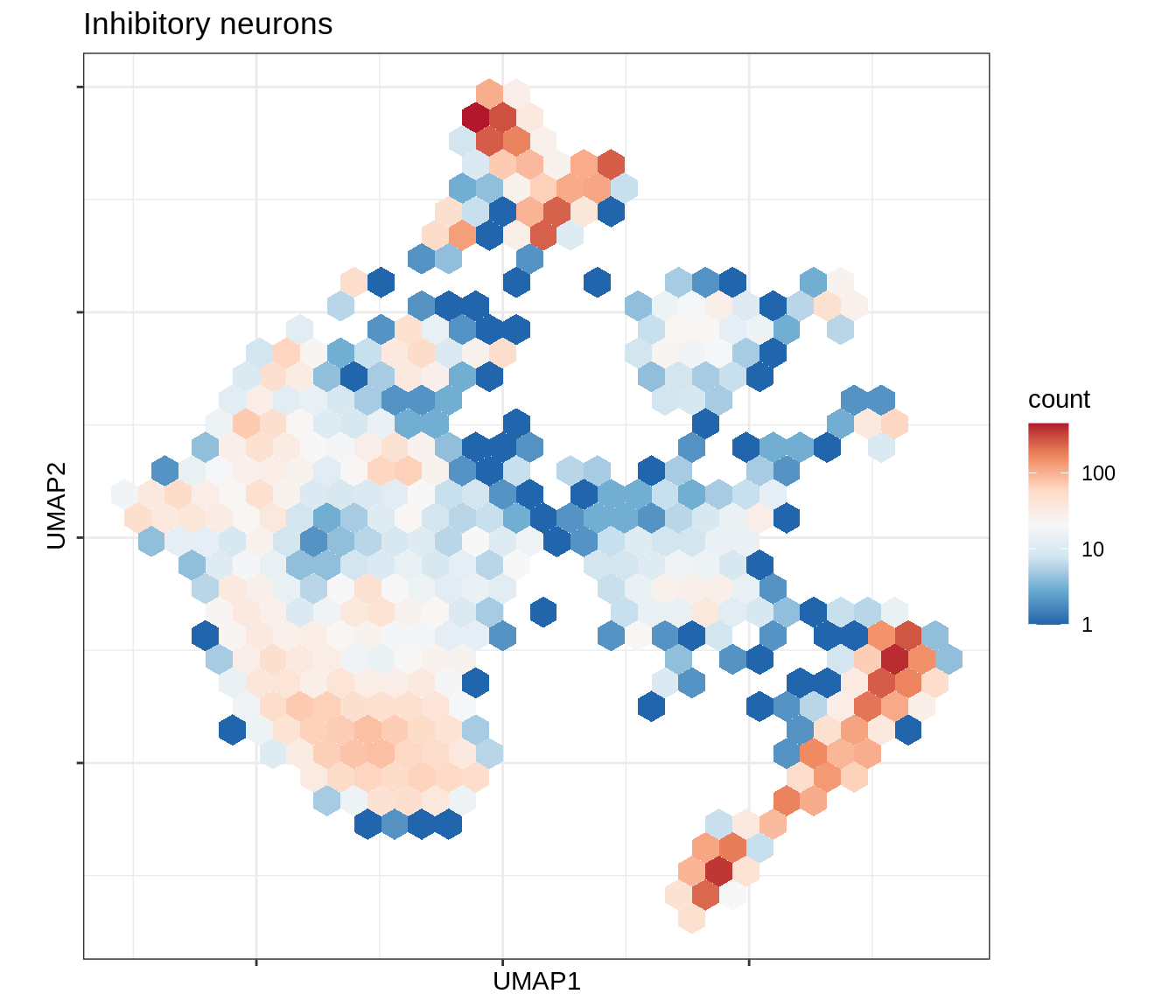
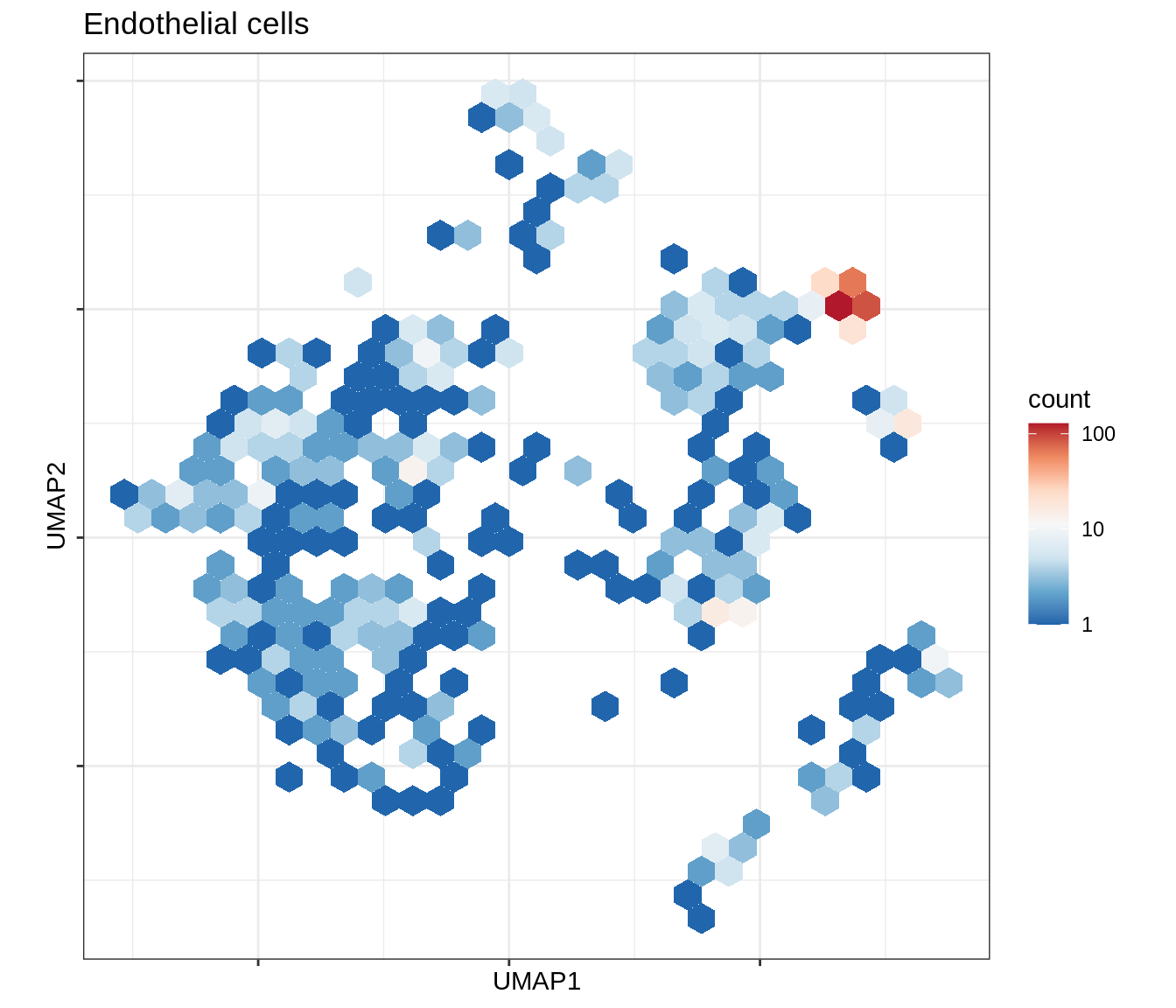
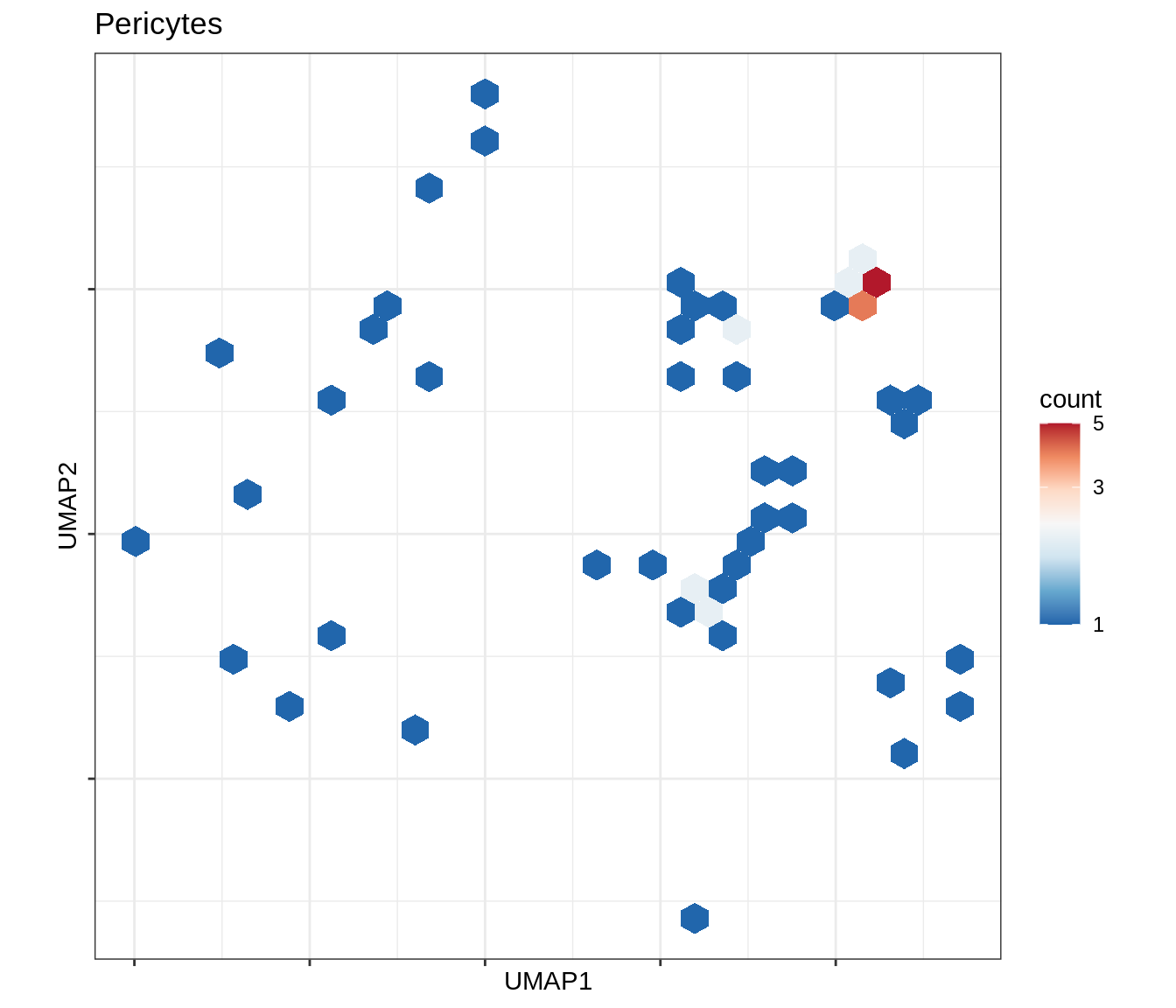
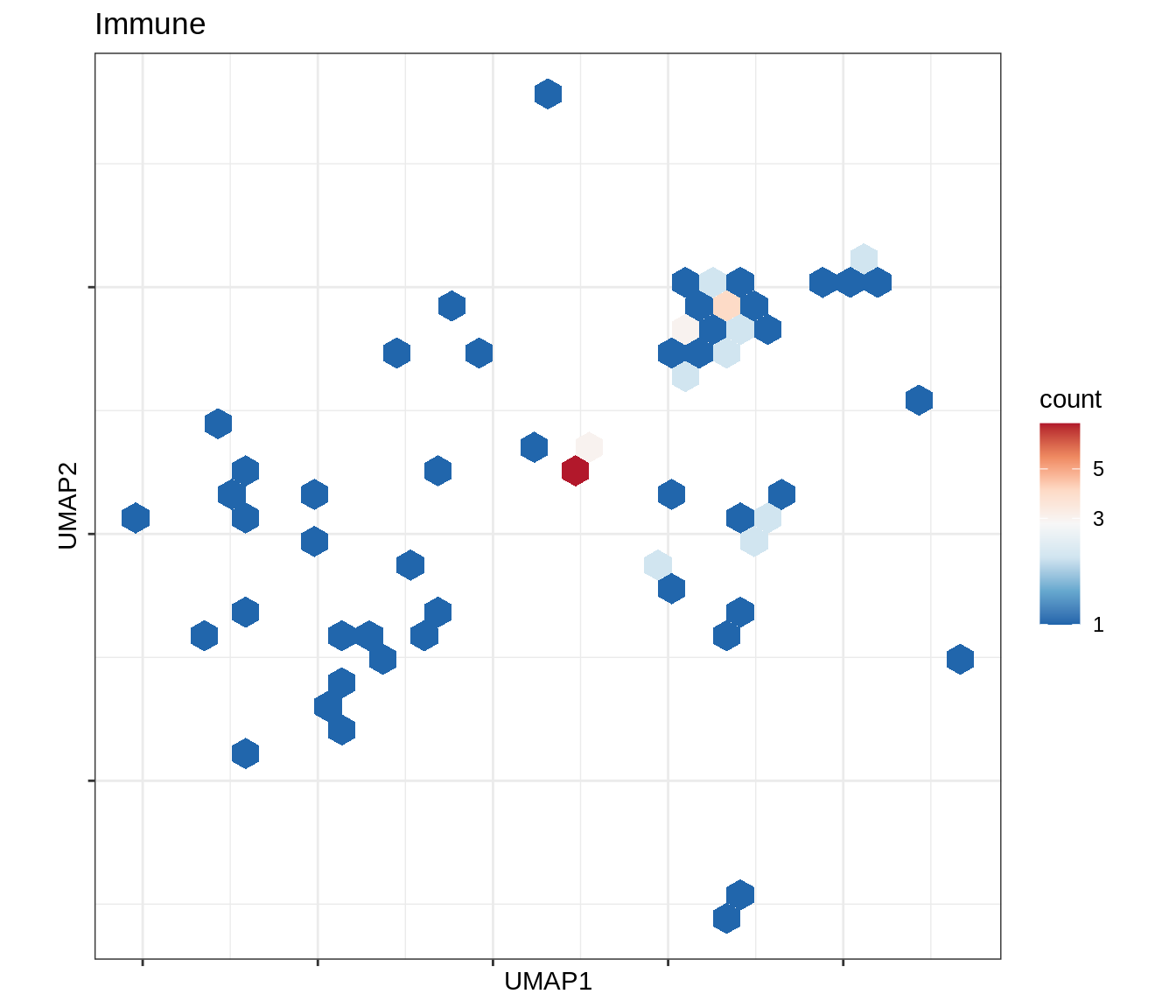
for (b in broad_ord) {
cat('### ', b, '\n')
print(plot_conos_density(conos_chk, conos_dt, b))
cat('\n\n')
}Outputs
devtools::session_info()- Session info ---------------------------------------------------------------
setting value
version R version 4.0.3 (2020-10-10)
os CentOS Linux 7 (Core)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_US.UTF-8
ctype C
tz Europe/Zurich
date 2021-07-28
- Packages -------------------------------------------------------------------
! package * version date lib
abind 1.4-5 2016-07-21 [2]
annotate 1.68.0 2020-10-27 [1]
AnnotationDbi 1.52.0 2020-10-27 [1]
assertthat * 0.2.1 2019-03-21 [2]
backports 1.2.1 2020-12-09 [2]
beachmat 2.6.4 2020-12-20 [1]
beeswarm 0.4.0 2021-06-01 [1]
Biobase * 2.50.0 2020-10-27 [1]
BiocGenerics * 0.36.1 2021-04-16 [1]
BiocManager 1.30.16 2021-06-15 [1]
BiocNeighbors 1.8.2 2020-12-07 [1]
BiocParallel * 1.24.1 2020-11-06 [1]
BiocSingular 1.6.0 2020-10-27 [1]
BiocStyle * 2.18.1 2020-11-24 [1]
Biostrings 2.58.0 2020-10-27 [1]
bit 4.0.4 2020-08-04 [2]
bit64 4.0.5 2020-08-30 [2]
bitops 1.0-7 2021-04-24 [2]
blme 1.0-5 2021-01-05 [1]
blob 1.2.1 2020-01-20 [2]
bluster 1.0.0 2020-10-27 [1]
boot 1.3-28 2021-05-03 [2]
brew 1.0-6 2011-04-13 [1]
broom 0.7.7 2021-06-13 [2]
bslib 0.2.5.1 2021-05-18 [2]
cachem 1.0.5 2021-05-15 [1]
Cairo 1.5-12.2 2020-07-07 [2]
callr 3.7.0 2021-04-20 [2]
caTools 1.18.2 2021-03-28 [2]
cellranger 1.1.0 2016-07-27 [2]
circlize * 0.4.13 2021-06-09 [1]
cli 2.5.0 2021-04-26 [2]
clue 0.3-59 2021-04-16 [1]
cluster 2.1.2 2021-04-17 [2]
codetools 0.2-18 2020-11-04 [2]
colorout * 1.2-2 2021-04-15 [1]
colorRamps 2.3 2012-10-29 [1]
colorspace 2.0-1 2021-05-04 [2]
ComplexHeatmap * 2.6.2 2020-11-12 [1]
conos * 1.4.1 2021-05-15 [1]
cowplot 1.1.1 2020-12-30 [2]
crayon 1.4.1 2021-02-08 [2]
data.table * 1.14.0 2021-02-21 [2]
DBI 1.1.1 2021-01-15 [2]
dbplyr 2.1.1 2021-04-06 [2]
DelayedArray 0.16.3 2021-03-24 [1]
DelayedMatrixStats 1.12.3 2021-02-03 [1]
deldir 0.2-10 2021-02-16 [2]
dendsort 0.3.4 2021-04-20 [1]
desc 1.3.0 2021-03-05 [2]
DESeq2 1.30.1 2021-02-19 [1]
devtools 2.4.2 2021-06-07 [1]
digest 0.6.27 2020-10-24 [2]
doParallel 1.0.16 2020-10-16 [1]
dplyr * 1.0.7 2021-06-18 [2]
dqrng 0.3.0 2021-05-01 [2]
drat 0.2.0 2021-04-22 [1]
DropletUtils * 1.10.3 2021-02-02 [1]
edgeR * 3.32.1 2021-01-14 [1]
ellipsis 0.3.2 2021-04-29 [2]
evaluate 0.14 2019-05-28 [2]
fansi 0.5.0 2021-05-25 [2]
farver 2.1.0 2021-02-28 [2]
fastmap 1.1.0 2021-01-25 [2]
fastmatch 1.1-0 2017-01-28 [1]
fgsea * 1.16.0 2020-10-27 [1]
fitdistrplus 1.1-5 2021-05-28 [2]
forcats * 0.5.1 2021-01-27 [2]
foreach 1.5.1 2020-10-15 [2]
fs 1.5.0 2020-07-31 [2]
future 1.21.0 2020-12-10 [2]
future.apply 1.7.0 2021-01-04 [2]
genefilter 1.72.1 2021-01-21 [1]
geneplotter 1.68.0 2020-10-27 [1]
generics 0.1.0 2020-10-31 [2]
GenomeInfoDb * 1.26.7 2021-04-08 [1]
GenomeInfoDbData 1.2.4 2021-04-15 [1]
GenomicAlignments 1.26.0 2020-10-27 [1]
GenomicRanges * 1.42.0 2020-10-27 [1]
GetoptLong 1.0.5 2020-12-15 [1]
ggbeeswarm * 0.6.0 2017-08-07 [1]
ggplot.multistats * 1.0.0 2019-10-28 [1]
ggplot2 * 3.3.4 2021-06-16 [1]
ggrepel * 0.9.1 2021-01-15 [2]
ggridges 0.5.3 2021-01-08 [2]
git2r 0.28.0 2021-01-10 [1]
glmmTMB 1.0.2.1 2020-07-02 [1]
GlobalOptions 0.1.2 2020-06-10 [1]
globals 0.14.0 2020-11-22 [2]
glue 1.4.2 2020-08-27 [2]
goftest 1.2-2 2019-12-02 [2]
googlesheets * 0.3.0 2018-06-29 [1]
gplots 3.1.1 2020-11-28 [2]
gridExtra 2.3 2017-09-09 [2]
grr 0.9.5 2016-08-26 [1]
gtable 0.3.0 2019-03-25 [2]
gtools 3.9.2 2021-06-06 [2]
haven 2.4.1 2021-04-23 [2]
HDF5Array 1.18.1 2021-02-04 [1]
hdf5r * 1.3.3 2020-08-18 [2]
hexbin 1.28.2 2021-01-08 [2]
highr 0.9 2021-04-16 [2]
hms 1.1.0 2021-05-17 [2]
htmltools 0.5.1.1 2021-01-22 [2]
htmlwidgets 1.5.3 2020-12-10 [2]
httpuv 1.6.1 2021-05-07 [2]
httr 1.4.2 2020-07-20 [2]
ica 1.0-2 2018-05-24 [2]
igraph * 1.2.6 2020-10-06 [2]
insight 0.14.2 2021-06-22 [1]
IRanges * 2.24.1 2020-12-12 [1]
irlba 2.3.3 2019-02-05 [2]
iterators * 1.0.13 2020-10-15 [2]
itertools * 0.1-3 2014-03-12 [1]
janitor 2.1.0 2021-01-05 [1]
jquerylib 0.1.4 2021-04-26 [2]
jsonlite 1.7.2 2020-12-09 [2]
kernlab 0.9-29 2019-11-12 [1]
KernSmooth 2.23-20 2021-05-03 [2]
knitr 1.33 2021-04-24 [1]
labeling 0.4.2 2020-10-20 [2]
later 1.2.0 2021-04-23 [2]
lattice 0.20-44 2021-05-02 [2]
lazyeval 0.2.2 2019-03-15 [2]
R leiden 0.3.8 <NA> [2]
leidenAlg 0.1.1 2021-03-03 [1]
lifecycle 1.0.0 2021-02-15 [2]
limma * 3.46.0 2020-10-27 [1]
listenv 0.8.0 2019-12-05 [2]
lme4 1.1-27.1 2021-06-22 [1]
lmerTest 3.1-3 2020-10-23 [1]
lmtest 0.9-38 2020-09-09 [2]
locfit 1.5-9.4 2020-03-25 [1]
loomR * 0.2.0 2021-04-15 [1]
lubridate 1.7.10 2021-02-26 [2]
magrittr * 2.0.1 2020-11-17 [1]
MASS 7.3-54 2021-05-03 [2]
Matrix * 1.3-4 2021-06-01 [2]
Matrix.utils * 0.9.8 2020-02-26 [1]
MatrixGenerics * 1.2.1 2021-01-30 [1]
matrixStats * 0.59.0 2021-06-01 [1]
mclust 5.4.7 2020-11-20 [1]
memoise 2.0.0 2021-01-26 [1]
mgcv 1.8-36 2021-06-01 [1]
mime 0.10 2021-02-13 [2]
miniUI 0.1.1.1 2018-05-18 [2]
minqa 1.2.4 2014-10-09 [1]
mixtools 1.2.0 2020-02-07 [1]
modelr 0.1.8 2020-05-19 [2]
munsell 0.5.0 2018-06-12 [2]
muscat * 1.5.1 2021-04-15 [1]
mvnfast 0.2.7 2021-05-20 [1]
mvtnorm 1.1-2 2021-06-07 [1]
N2R 0.1.1 2020-12-14 [1]
nlme 3.1-152 2021-02-04 [2]
nloptr 1.2.2.2 2020-07-02 [1]
nnls * 1.4 2012-03-19 [1]
numDeriv 2016.8-1.1 2019-06-06 [2]
pagoda2 * 1.0.3 2021-05-03 [1]
parallelly 1.26.0 2021-06-09 [2]
patchwork * 1.1.1 2020-12-17 [2]
pbapply 1.4-3 2020-08-18 [2]
pbkrtest 0.5.1 2021-03-09 [1]
performance * 0.7.2 2021-05-17 [1]
pillar 1.6.1 2021-05-16 [2]
pkgbuild 1.2.0 2020-12-15 [1]
pkgconfig 2.0.3 2019-09-22 [2]
pkgload 1.2.1 2021-04-06 [2]
plotly 4.9.3 2021-01-10 [2]
plyr 1.8.6 2020-03-03 [2]
png 0.1-7 2013-12-03 [2]
polyclip 1.10-0 2019-03-14 [2]
prettyunits 1.1.1 2020-01-24 [2]
processx 3.5.2 2021-04-30 [2]
progress 1.2.2 2019-05-16 [2]
promises 1.2.0.1 2021-02-11 [2]
ps 1.6.0 2021-02-28 [2]
purrr * 0.3.4 2020-04-17 [2]
R.methodsS3 1.8.1 2020-08-26 [1]
R.oo 1.24.0 2020-08-26 [1]
R.utils 2.10.1 2020-08-26 [1]
R6 * 2.5.0 2020-10-28 [2]
ranger * 0.12.1 2020-01-10 [1]
RANN 2.6.1 2019-01-08 [2]
RColorBrewer * 1.1-2 2014-12-07 [2]
Rcpp 1.0.6 2021-01-15 [2]
RcppAnnoy 0.0.18 2020-12-15 [2]
RCurl 1.98-1.3 2021-03-16 [1]
readr * 1.4.0 2020-10-05 [2]
readxl * 1.3.1 2019-03-13 [2]
registry 0.5-1 2019-03-05 [1]
remotes 2.4.0 2021-06-02 [1]
reprex 2.0.0 2021-04-02 [2]
reshape2 * 1.4.4 2020-04-09 [2]
reticulate 1.20 2021-05-03 [2]
rhdf5 2.34.0 2020-10-27 [1]
rhdf5filters 1.2.1 2021-05-03 [1]
Rhdf5lib 1.12.1 2021-01-26 [1]
rjson 0.2.20 2018-06-08 [1]
rlang 0.4.11 2021-04-30 [2]
rmarkdown * 2.8 2021-05-07 [2]
RMTstat 0.3 2014-11-01 [1]
ROCR 1.0-11 2020-05-02 [2]
Rook 1.1-1 2014-10-20 [1]
rpart 4.1-15 2019-04-12 [2]
rprojroot 2.0.2 2020-11-15 [2]
Rsamtools 2.6.0 2020-10-27 [1]
RSQLite 2.2.7 2021-04-22 [1]
rstudioapi 0.13 2020-11-12 [2]
rsvd 1.0.5 2021-04-16 [1]
rtracklayer * 1.50.0 2020-10-27 [1]
Rtsne 0.15 2018-11-10 [2]
rvest 1.0.0 2021-03-09 [2]
S4Vectors * 0.28.1 2020-12-09 [1]
SampleQC * 0.4.5 2021-04-15 [1]
sass 0.4.0 2021-05-12 [2]
scales * 1.1.1 2020-05-11 [2]
scater * 1.18.6 2021-02-26 [1]
scattermore 0.7 2020-11-24 [2]
sccore 0.1.3 2021-05-05 [1]
scran * 1.18.7 2021-04-16 [1]
sctransform 0.3.2 2020-12-16 [2]
scuttle 1.0.4 2020-12-17 [1]
segmented 1.3-4 2021-04-22 [1]
seriation * 1.2-9 2020-10-01 [1]
sessioninfo 1.1.1 2018-11-05 [1]
Seurat * 4.0.1 2021-03-18 [2]
SeuratObject * 4.0.1 2021-05-08 [2]
shape 1.4.6 2021-05-19 [1]
shiny 1.6.0 2021-01-25 [2]
SingleCellExperiment * 1.12.0 2020-10-27 [1]
snakecase 0.11.0 2019-05-25 [1]
sparseMatrixStats 1.2.1 2021-02-02 [1]
spatstat.core 2.1-2 2021-04-18 [2]
spatstat.data 2.1-0 2021-03-21 [2]
spatstat.geom 2.1-0 2021-04-15 [2]
spatstat.sparse 2.0-0 2021-03-16 [2]
spatstat.utils 2.2-0 2021-06-14 [2]
statmod 1.4.36 2021-05-10 [1]
stringi 1.6.2 2021-05-17 [2]
stringr * 1.4.0 2019-02-10 [2]
SummarizedExperiment * 1.20.0 2020-10-27 [1]
survival 3.2-11 2021-04-26 [2]
tensor 1.5 2012-05-05 [2]
testthat 3.0.3 2021-06-16 [2]
tibble * 3.1.2 2021-05-16 [2]
tictoc * 1.0.1 2021-04-19 [1]
tidyr * 1.1.3 2021-03-03 [2]
tidyselect 1.1.1 2021-04-30 [2]
tidyverse * 1.3.1 2021-04-15 [2]
TMB 1.7.20 2021-04-08 [1]
triebeard 0.3.0 2016-08-04 [2]
TSP 1.1-10 2020-04-17 [1]
UpSetR * 1.4.0 2019-05-22 [1]
urltools 1.7.3 2019-04-14 [2]
usethis 2.0.1 2021-02-10 [1]
utf8 1.2.1 2021-03-12 [2]
uwot * 0.1.10 2020-12-15 [2]
variancePartition 1.20.0 2020-10-27 [1]
vctrs 0.3.8 2021-04-29 [2]
vipor 0.4.5 2017-03-22 [1]
viridis * 0.6.1 2021-05-11 [1]
viridisLite * 0.4.0 2021-04-13 [1]
whisker 0.4 2019-08-28 [1]
withr 2.4.2 2021-04-18 [2]
workflowr * 1.6.2 2020-04-30 [1]
writexl * 1.4.0 2021-04-20 [1]
xfun 0.24 2021-06-15 [1]
XML 3.99-0.6 2021-03-16 [1]
xml2 1.3.2 2020-04-23 [2]
xtable 1.8-4 2019-04-21 [2]
XVector 0.30.0 2020-10-27 [1]
yaml 2.2.1 2020-02-01 [2]
zlibbioc 1.36.0 2020-10-27 [1]
zoo 1.8-9 2021-03-09 [2]
source
CRAN (R 4.0.0)
Bioconductor
Bioconductor
CRAN (R 4.0.0)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
Bioconductor
Bioconductor
CRAN (R 4.0.3)
Bioconductor
Bioconductor
Bioconductor
Bioconductor
Bioconductor
CRAN (R 4.0.2)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.1)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Github (jalvesaq/colorout@79931fd)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
Bioconductor
CRAN (R 4.0.3)
Bioconductor
Bioconductor
Bioconductor
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.2)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
<NA>
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Github (mojaveazure/loomR@df0144b)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.1)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
Github (HelenaLC/muscat@c939663)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
Bioconductor
Bioconductor
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.0)
CRAN (R 4.0.3)
Bioconductor
Github (wmacnair/SampleQC@5e3f021)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.1)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.1)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
[1] /pstore/home/macnairw/lib/conda_r3.12
[2] /pstore/home/macnairw/.conda/envs/r_4.0.3/lib/R/library
R -- Package was removed from disk.Figures
Dotplots of muscat genes marker genes (all samples)
top_muscat = muscat_all[ logFC > 0 ] %>%
.[, fdr_rank := frank(FDR, ties.method = 'random'), by = type_fine ] %>%
.[ fdr_rank <= 2 ] %>%
.[ order(type_broad, type_fine, fdr_rank) ] %>%
.[, .(broad_class = type_broad, symbol) ]
(plot_dotplot(pb_fine, props_fine, top_muscat, labels_dt,
row_split = 'broad_class'))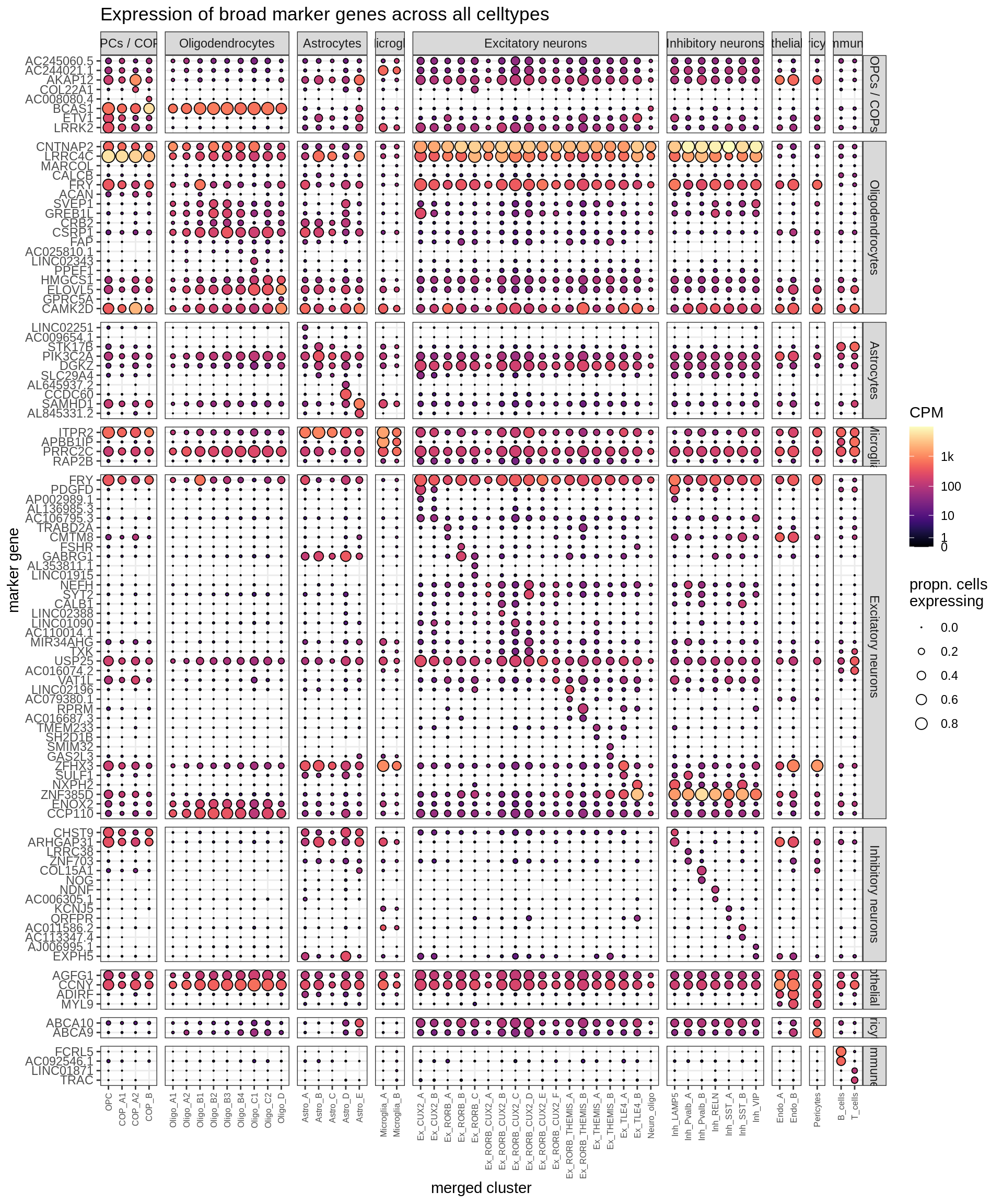
Dotplots of muscat genes marker genes (healthy samples)
top_muscat = muscat_ctrl[ logFC > 0 ] %>%
.[, fdr_rank := frank(FDR, ties.method = 'random'), by = type_fine ] %>%
.[ fdr_rank <= 2 ] %>%
.[ order(type_broad, type_fine, fdr_rank) ] %>%
.[, .(broad_class = type_broad, symbol) ]
(plot_dotplot(pb_fine, props_fine, top_muscat, labels_dt,
row_split = 'broad_class'))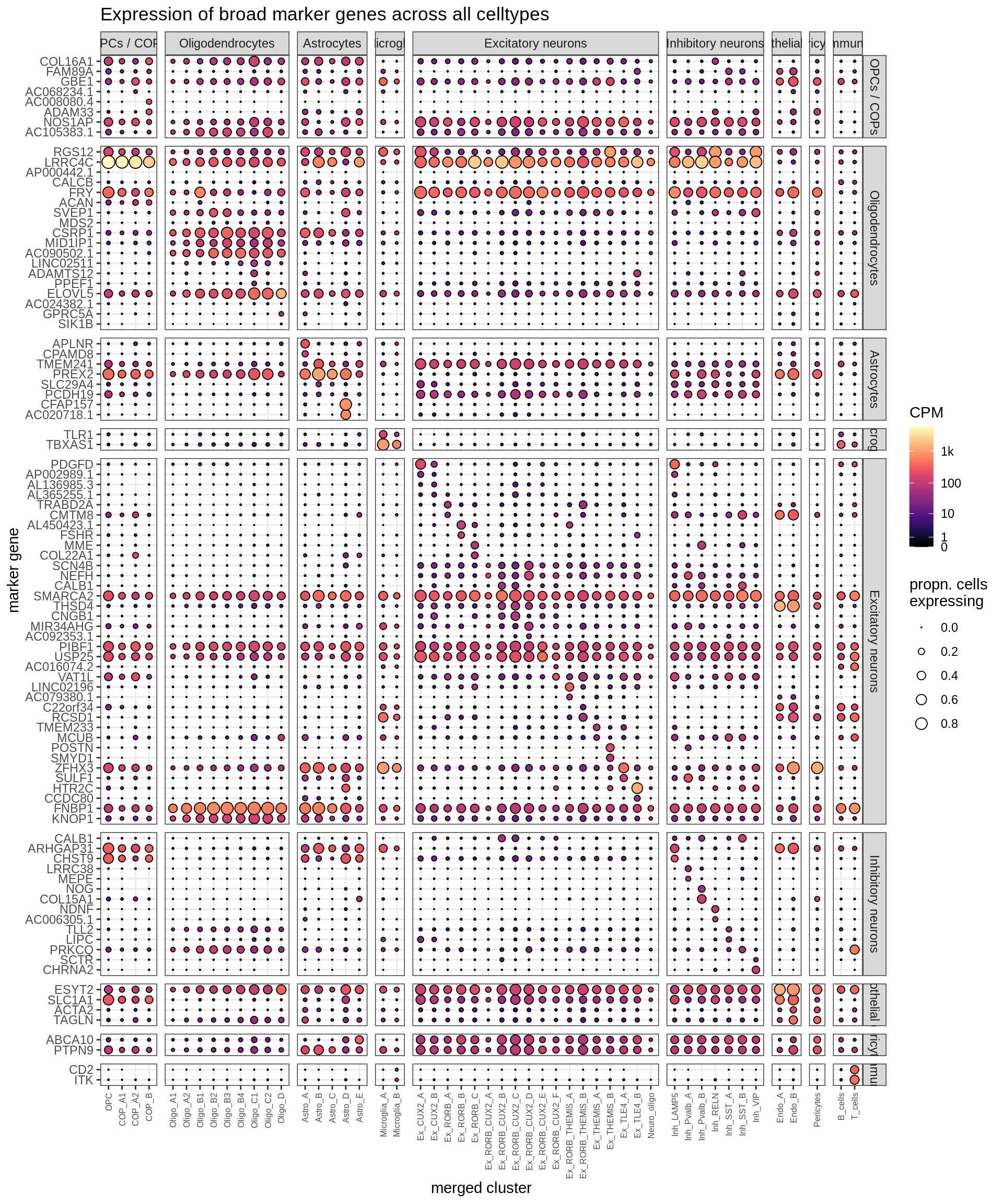
Dotplots of known oligo genes
oligo_gs = list(
oligos = c("MAG", "MOG", "CNP", "MYRF", "PLP1", "MBP"),
lipids = c("ELOVL2", "ELOVL4", "ELOVL5", "ELOVL7"),
myelin = c("CERS2", "CGT", "CST", "DAPAT", "PEX7")
)
oligos_dt = lapply(names(oligo_gs), function(n)
data.table(oligo_fn = n, symbol = oligo_gs[[n]]) ) %>% rbindlist %>%
.[, oligo_fn := factor(oligo_fn, levels = names(oligo_gs))]
olg_types = c("Oligo_A1", "Oligo_A2", "Oligo_B1", "Oligo_B2", "Oligo_B3", "Oligo_B4",
"Oligo_C1", "Oligo_C2", "Oligo_D")
pb_olgs = copy(pb_fine)
assays(pb_olgs) = assays(pb_olgs)[olg_types]
int_colData(pb_olgs)$n_cells =
lapply(int_colData(pb_olgs)$n_cells, function(l) l[olg_types])
props_olgs = copy(props_fine)
assays(props_olgs) = assays(props_olgs)[olg_types]
int_colData(props_olgs)$n_cells =
lapply(int_colData(props_olgs)$n_cells, function(l) l[olg_types])
(plot_dotplot(pb_olgs, props_olgs, oligos_dt, labels_dt,
row_split = 'oligo_fn'))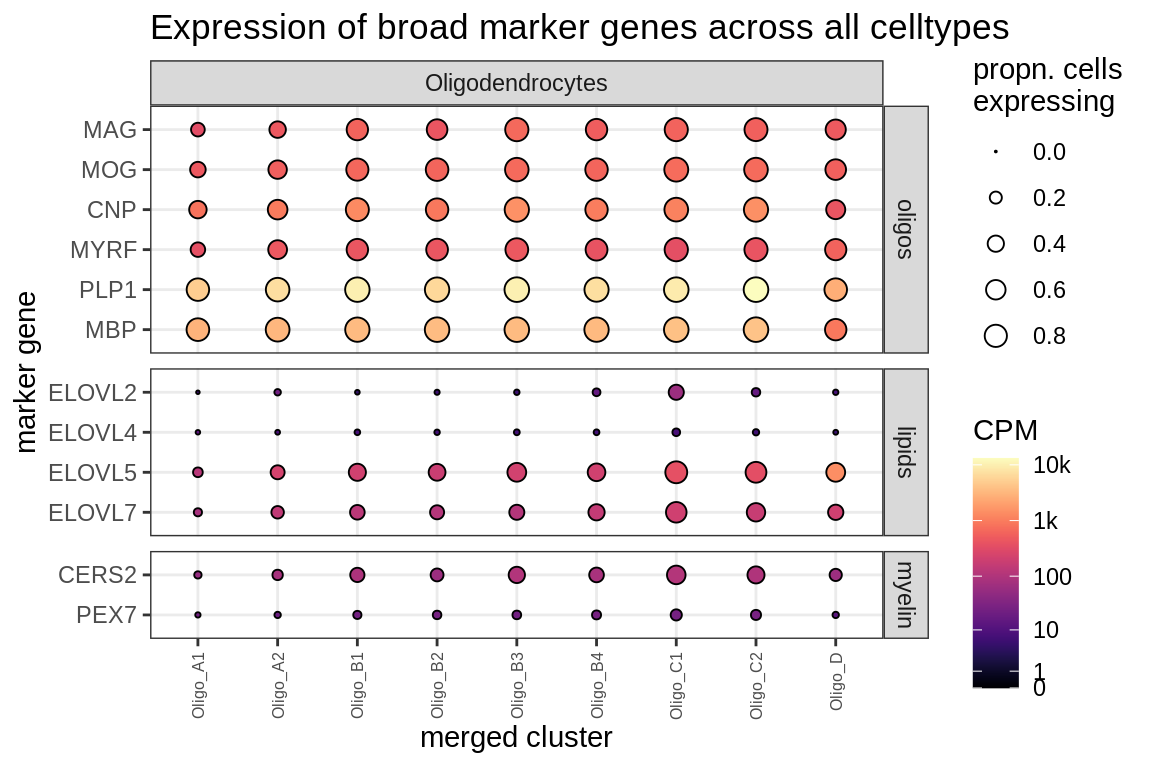
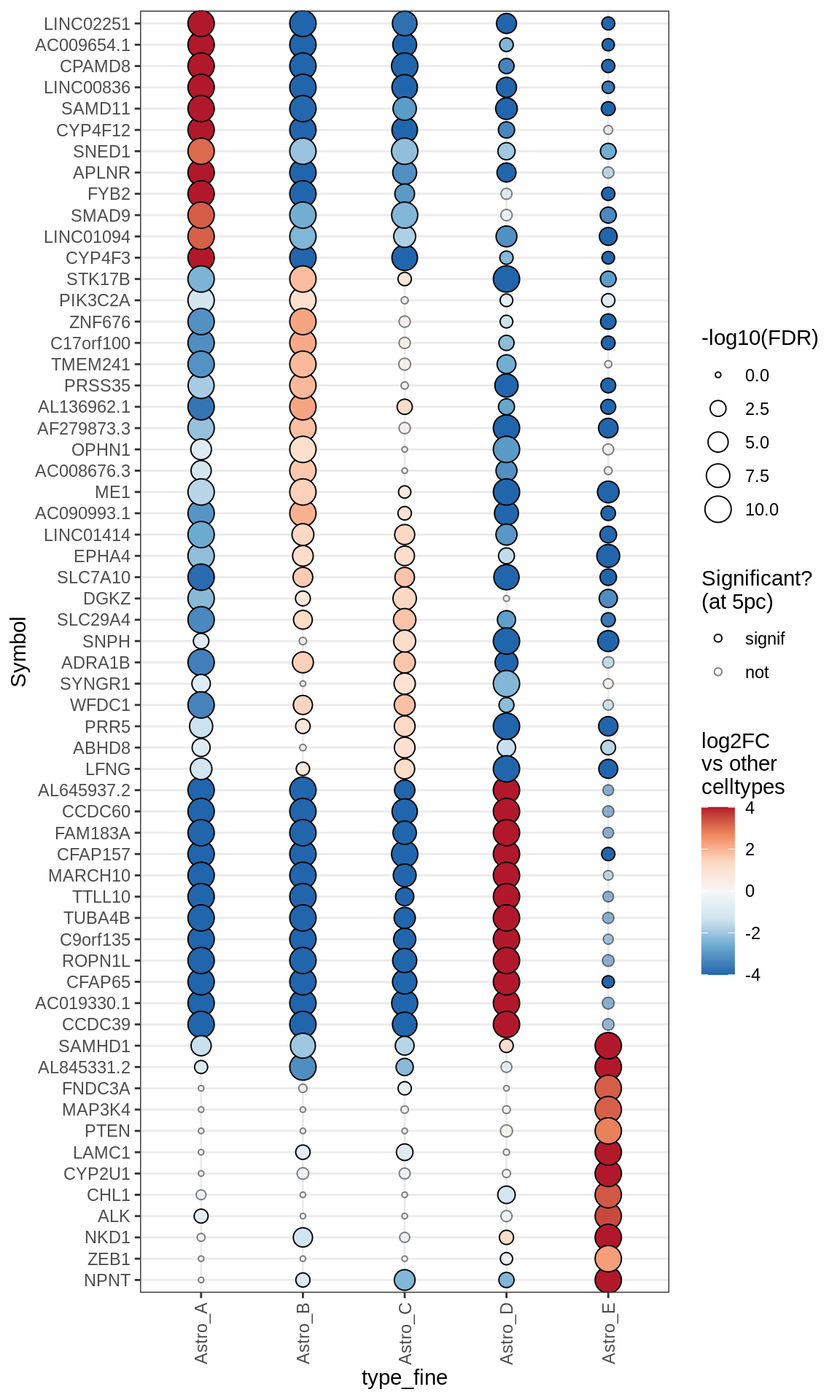
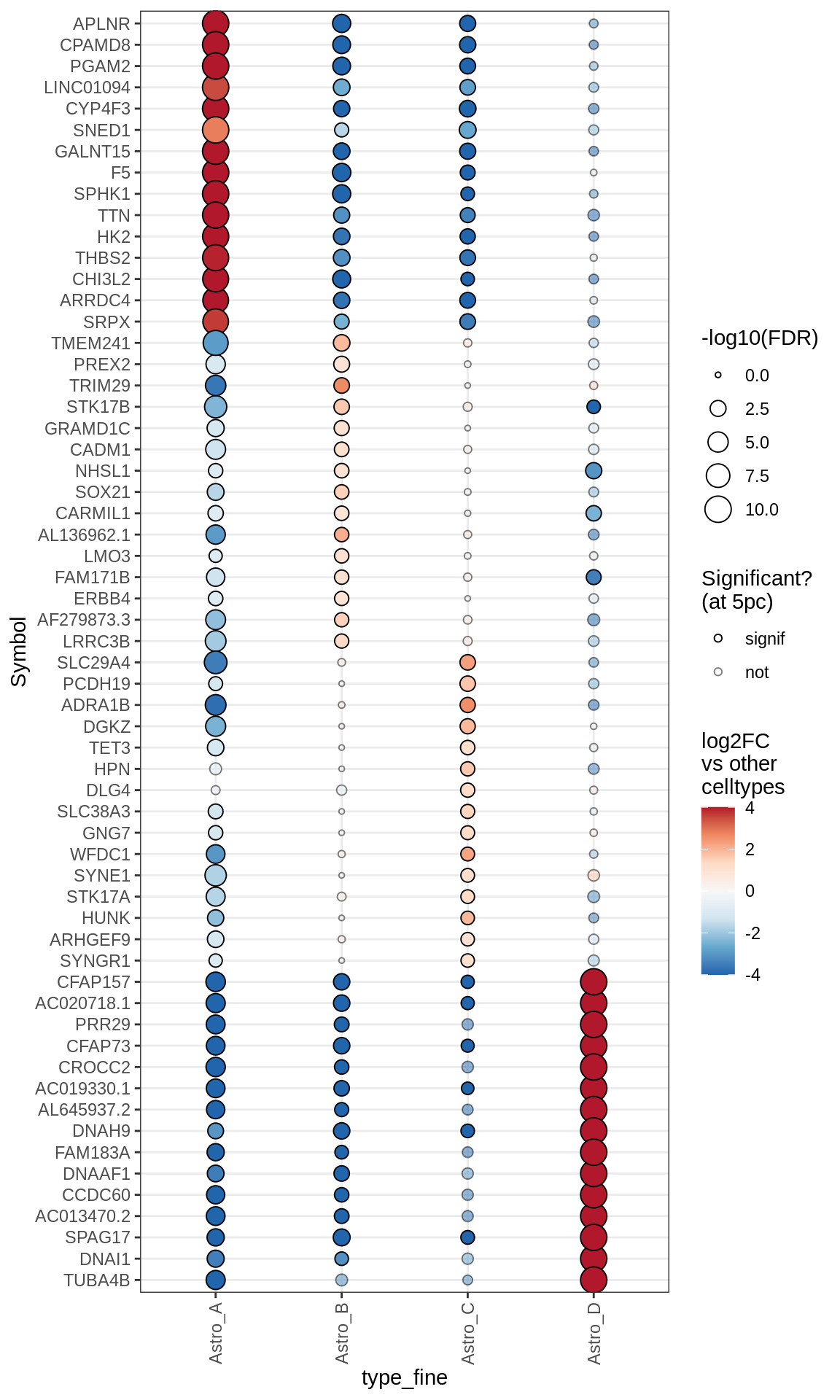
Dotplots of astrocyte layer genes
astro_gs = list(
pan_astro = c("ALDH1A1", "GFAP"),
upper = c("SCEL", "SPRY1", "DDHD1", "EOGT", "CHRDL1", "ADIPOR2"),
gm_bias = c("GRM3", "KIRREL2", "IGFBP2", "IRAK2", "TRPM3", "CHD9",
"INKA2", "SLC25A34", "AXIN2", "SLC7A5", "LIMD1", "CDO1", "AKT2", "LEF1",
"BSG", "APOE", "MFGE8", "ITM2C", "SLC27A1", "CYP4F2", "FJX1"),
deep = c("ID3", "ID1", "DKK3", "EFHD2", "LRP1B", "DHCR24",
"IL33", "PAQR6")
)
astros_dt = lapply(names(astro_gs), function(n)
data.table(astro_layer = n, symbol = astro_gs[[n]]) ) %>% rbindlist %>%
.[, astro_layer := factor(astro_layer, levels = names(astro_gs))]
# setdiff(astros_dt$symbol, rowData(pb_fine)$symbol)
ast_types = c("Astro_A", "Astro_B", "Astro_C", "Astro_D", "Astro_E")
pb_asts = copy(pb_fine)
assays(pb_asts) = assays(pb_asts)[ast_types]
int_colData(pb_asts)$n_cells =
lapply(int_colData(pb_asts)$n_cells, function(l) l[ast_types])
props_asts = copy(props_fine)
assays(props_asts) = assays(props_asts)[ast_types]
int_colData(props_asts)$n_cells =
lapply(int_colData(props_asts)$n_cells, function(l) l[ast_types])
(plot_dotplot(pb_asts, props_asts, astros_dt, labels_dt,
row_split = 'astro_layer'))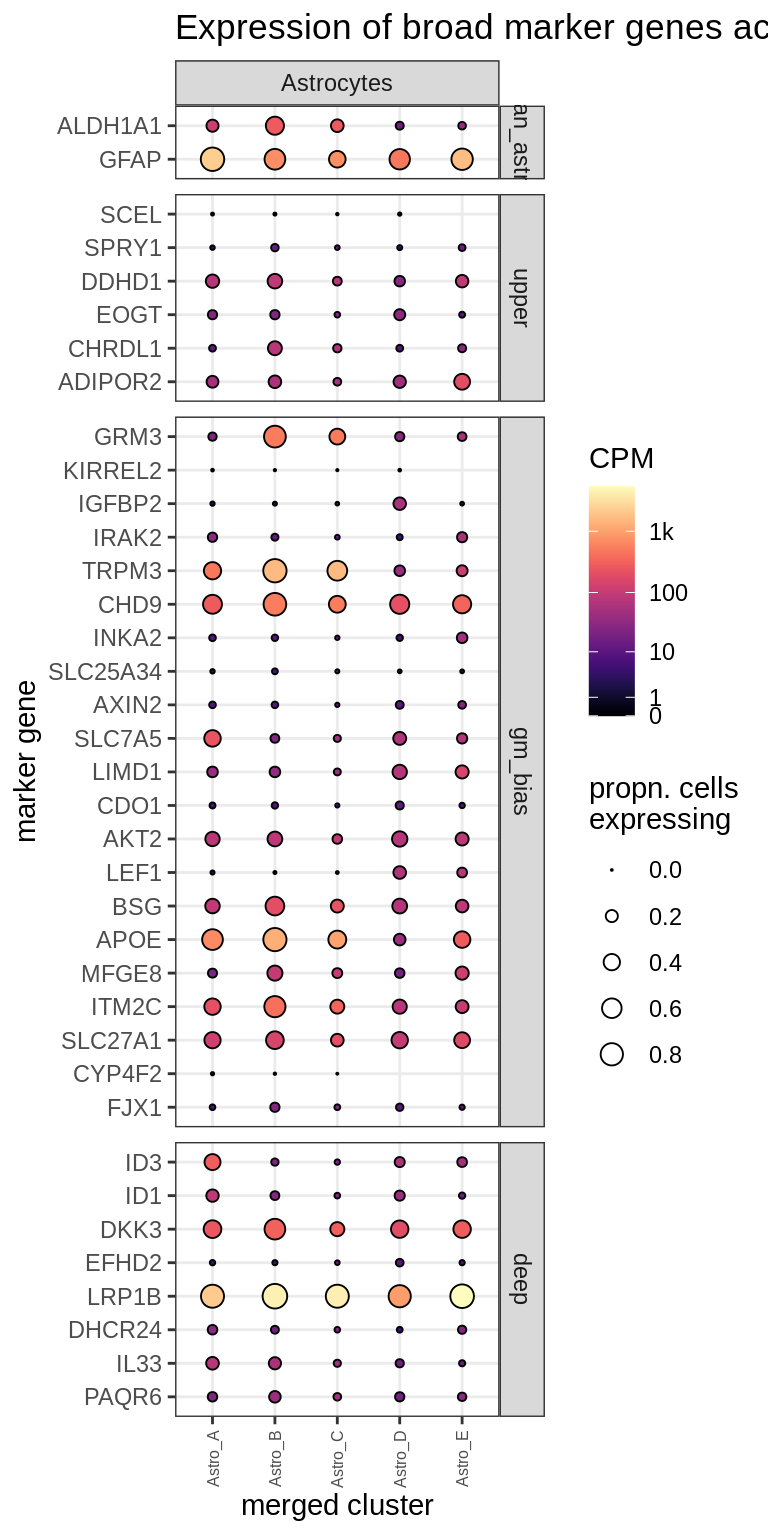
| Version | Author | Date |
|---|---|---|
| f60e5c1 | Macnair | 2021-08-03 |
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-conda-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /pstore/home/macnairw/.conda/envs/r_4.0.3/lib/libopenblasp-r0.3.12.so
locale:
[1] LC_CTYPE=C LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] grid parallel stats4 stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] ggbeeswarm_0.6.0 pagoda2_1.0.3
[3] ranger_0.12.1 rmarkdown_2.8
[5] ggrepel_0.9.1 writexl_1.4.0
[7] readxl_1.3.1 fgsea_1.16.0
[9] tictoc_1.0.1 performance_0.7.2
[11] reshape2_1.4.4 Matrix.utils_0.9.8
[13] UpSetR_1.4.0 dplyr_1.0.7
[15] purrr_0.3.4 readr_1.4.0
[17] tidyr_1.1.3 tibble_3.1.2
[19] tidyverse_1.3.1 rtracklayer_1.50.0
[21] nnls_1.4 muscat_1.5.1
[23] DropletUtils_1.10.3 edgeR_3.32.1
[25] limma_3.46.0 googlesheets_0.3.0
[27] scran_1.18.7 uwot_0.1.10
[29] scater_1.18.6 BiocParallel_1.24.1
[31] ggplot.multistats_1.0.0 seriation_1.2-9
[33] ComplexHeatmap_2.6.2 SeuratObject_4.0.1
[35] Seurat_4.0.1 conos_1.4.1
[37] igraph_1.2.6 SampleQC_0.4.5
[39] SingleCellExperiment_1.12.0 SummarizedExperiment_1.20.0
[41] Biobase_2.50.0 GenomicRanges_1.42.0
[43] GenomeInfoDb_1.26.7 IRanges_2.24.1
[45] S4Vectors_0.28.1 BiocGenerics_0.36.1
[47] MatrixGenerics_1.2.1 matrixStats_0.59.0
[49] Matrix_1.3-4 loomR_0.2.0
[51] itertools_0.1-3 iterators_1.0.13
[53] hdf5r_1.3.3 R6_2.5.0
[55] patchwork_1.1.1 forcats_0.5.1
[57] ggplot2_3.3.4 scales_1.1.1
[59] viridis_0.6.1 viridisLite_0.4.0
[61] assertthat_0.2.1 stringr_1.4.0
[63] data.table_1.14.0 magrittr_2.0.1
[65] circlize_0.4.13 RColorBrewer_1.1-2
[67] BiocStyle_2.18.1 colorout_1.2-2
[69] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] rsvd_1.0.5 ica_1.0-2
[3] ps_1.6.0 Rsamtools_2.6.0
[5] foreach_1.5.1 lmtest_0.9-38
[7] rprojroot_2.0.2 crayon_1.4.1
[9] spatstat.core_2.1-2 MASS_7.3-54
[11] rhdf5filters_1.2.1 nlme_3.1-152
[13] backports_1.2.1 reprex_2.0.0
[15] rlang_0.4.11 XVector_0.30.0
[17] ROCR_1.0-11 irlba_2.3.3
[19] callr_3.7.0 nloptr_1.2.2.2
[21] rjson_0.2.20 bit64_4.0.5
[23] glue_1.4.2 sctransform_0.3.2
[25] processx_3.5.2 pbkrtest_0.5.1
[27] vipor_0.4.5 spatstat.sparse_2.0-0
[29] AnnotationDbi_1.52.0 spatstat.geom_2.1-0
[31] haven_2.4.1 tidyselect_1.1.1
[33] usethis_2.0.1 fitdistrplus_1.1-5
[35] variancePartition_1.20.0 XML_3.99-0.6
[37] zoo_1.8-9 GenomicAlignments_1.26.0
[39] xtable_1.8-4 evaluate_0.14
[41] cli_2.5.0 scuttle_1.0.4
[43] zlibbioc_1.36.0 rstudioapi_0.13
[45] miniUI_0.1.1.1 whisker_0.4
[47] bslib_0.2.5.1 rpart_4.1-15
[49] fastmatch_1.1-0 shiny_1.6.0
[51] BiocSingular_1.6.0 xfun_0.24
[53] clue_0.3-59 pkgbuild_1.2.0
[55] cluster_2.1.2 caTools_1.18.2
[57] TSP_1.1-10 listenv_0.8.0
[59] Biostrings_2.58.0 png_0.1-7
[61] future_1.21.0 withr_2.4.2
[63] bitops_1.0-7 plyr_1.8.6
[65] cellranger_1.1.0 dqrng_0.3.0
[67] pillar_1.6.1 gplots_3.1.1
[69] GlobalOptions_0.1.2 cachem_1.0.5
[71] fs_1.5.0 kernlab_0.9-29
[73] GetoptLong_1.0.5 DelayedMatrixStats_1.12.3
[75] vctrs_0.3.8 ellipsis_0.3.2
[77] generics_0.1.0 devtools_2.4.2
[79] urltools_1.7.3 tools_4.0.3
[81] beeswarm_0.4.0 munsell_0.5.0
[83] DelayedArray_0.16.3 pkgload_1.2.1
[85] fastmap_1.1.0 compiler_4.0.3
[87] abind_1.4-5 httpuv_1.6.1
[89] segmented_1.3-4 sessioninfo_1.1.1
[91] plotly_4.9.3 GenomeInfoDbData_1.2.4
[93] gridExtra_2.3 glmmTMB_1.0.2.1
[95] lattice_0.20-44 deldir_0.2-10
[97] utf8_1.2.1 later_1.2.0
[99] jsonlite_1.7.2 dendsort_0.3.4
[101] pbapply_1.4-3 sparseMatrixStats_1.2.1
[103] genefilter_1.72.1 lazyeval_0.2.2
[105] promises_1.2.0.1 doParallel_1.0.16
[107] R.utils_2.10.1 goftest_1.2-2
[109] brew_1.0-6 spatstat.utils_2.2-0
[111] reticulate_1.20 cowplot_1.1.1
[113] blme_1.0-5 statmod_1.4.36
[115] Rtsne_0.15 Rook_1.1-1
[117] HDF5Array_1.18.1 survival_3.2-11
[119] numDeriv_2016.8-1.1 yaml_2.2.1
[121] htmltools_0.5.1.1 memoise_2.0.0
[123] locfit_1.5-9.4 digest_0.6.27
[125] mime_0.10 registry_0.5-1
[127] N2R_0.1.1 RSQLite_2.2.7
[129] future.apply_1.7.0 remotes_2.4.0
[131] blob_1.2.1 R.oo_1.24.0
[133] drat_0.2.0 mvnfast_0.2.7
[135] labeling_0.4.2 splines_4.0.3
[137] Rhdf5lib_1.12.1 Cairo_1.5-12.2
[139] mixtools_1.2.0 RCurl_1.98-1.3
[141] broom_0.7.7 hms_1.1.0
[143] modelr_0.1.8 rhdf5_2.34.0
[145] colorspace_2.0-1 BiocManager_1.30.16
[147] shape_1.4.6 sass_0.4.0
[149] Rcpp_1.0.6 mclust_5.4.7
[151] RANN_2.6.1 mvtnorm_1.1-2
[153] fansi_0.5.0 parallelly_1.26.0
[155] ggridges_0.5.3 lifecycle_1.0.0
[157] bluster_1.0.0 minqa_1.2.4
[159] testthat_3.0.3 leiden_0.3.8
[161] jquerylib_0.1.4 snakecase_0.11.0
[163] desc_1.3.0 RcppAnnoy_0.0.18
[165] TMB_1.7.20 htmlwidgets_1.5.3
[167] triebeard_0.3.0 RMTstat_0.3
[169] beachmat_2.6.4 polyclip_1.10-0
[171] rvest_1.0.0 mgcv_1.8-36
[173] globals_0.14.0 insight_0.14.2
[175] leidenAlg_0.1.1 lubridate_1.7.10
[177] codetools_0.2-18 gtools_3.9.2
[179] prettyunits_1.1.1 dbplyr_2.1.1
[181] R.methodsS3_1.8.1 gtable_0.3.0
[183] DBI_1.1.1 git2r_0.28.0
[185] tensor_1.5 httr_1.4.2
[187] highr_0.9 KernSmooth_2.23-20
[189] stringi_1.6.2 progress_1.2.2
[191] farver_2.1.0 annotate_1.68.0
[193] hexbin_1.28.2 xml2_1.3.2
[195] colorRamps_2.3 sccore_0.1.3
[197] boot_1.3-28 grr_0.9.5
[199] BiocNeighbors_1.8.2 lme4_1.1-27.1
[201] geneplotter_1.68.0 scattermore_0.7
[203] DESeq2_1.30.1 bit_4.0.4
[205] spatstat.data_2.1-0 janitor_2.1.0
[207] pkgconfig_2.0.3 lmerTest_3.1-3
[209] knitr_1.33