MS: Identifying contamination by ambient RNA
Will Macnair
Institute for Molecular Life Sciences, University of Zurich, SwitzerlandSwiss Institute of Bioinformatics (SIB), University of Zurich, SwitzerlandOctober 26, 2021
Last updated: 2021-10-26
Checks: 4 3
Knit directory: MS_lesions/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
The global environment had objects present when the code in the R Markdown file was run. These objects can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment. Use wflow_publish or wflow_build to ensure that the code is always run in an empty environment.
The following objects were defined in the global environment when these results were created:
| Name | Class | Size |
|---|---|---|
| q | function | 1008 bytes |
The command set.seed(20210118) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
- calc_contamination
- calc_contamination_negative_control
- calc_find_markers_soup
- calc_pseudobulk
- calc_soup_profile
- calc_soup_props
- check_contamination_broad
- check_contamination_fine
- load_conos
- load_gene_cats
- load_metadata
- load_soup
- load_top_markers
- plot_dotplot_findmarkers
- plot_soup_max_vs_ctrl
- plot_soup_props_by_cluster
- plot_soup_props_vs_qc_celltype
- plot_soup_props_vs_qc_max
- plot_soup_props_vs_qc_mt
- plot_soup_props_vs_qc_split_by_celltype
- save_outputs
- session_info
- session-info-chunk-inserted-by-workflowr
- setup_input
- setup_outputs
To ensure reproducibility of the results, delete the cache directory ms07_soup_cache and re-run the analysis. To have workflowr automatically delete the cache directory prior to building the file, set delete_cache = TRUE when running wflow_build() or wflow_publish().
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version b9e43a8. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rprofile
Ignored: .Rproj.user/
Ignored: ._MS_lesions.sublime-project
Ignored: .log/
Ignored: MS_lesions.sublime-project
Ignored: MS_lesions.sublime-workspace
Ignored: analysis/.__site.yml
Ignored: analysis/fig_muscat_cache/
Ignored: analysis/ms02_doublet_id_cache/
Ignored: analysis/ms03_SampleQC_cache/
Ignored: analysis/ms04_conos_cache/
Ignored: analysis/ms05_splitting_cache/
Ignored: analysis/ms06_sccaf_cache/
Ignored: analysis/ms07_soup_cache/
Ignored: analysis/ms08_modules_cache/
Ignored: analysis/ms08_modules_pseudobulk_cache/
Ignored: analysis/ms09_ancombc_cache/
Ignored: analysis/ms09_ancombc_clean_1e3_cache/
Ignored: analysis/ms09_ancombc_clean_2e3_cache/
Ignored: analysis/ms09_ancombc_mixed_cache/
Ignored: analysis/ms10_muscat_run01_cache/
Ignored: analysis/ms10_muscat_run02_cache/
Ignored: analysis/ms10_muscat_template_broad_cache/
Ignored: analysis/ms10_muscat_template_fine_cache/
Ignored: analysis/ms11_paga_cache/
Ignored: analysis/ms12_markers_cache/
Ignored: analysis/ms13_labelling_cache/
Ignored: analysis/ms14_lesions_cache/
Ignored: analysis/ms15_mofa_sample_gm_cache/
Ignored: analysis/ms15_mofa_sample_gm_final_meta_cache/
Ignored: analysis/ms15_mofa_sample_gm_superclean_cache/
Ignored: analysis/ms15_mofa_sample_wm_cache/
Ignored: analysis/ms15_mofa_sample_wm_final_meta_cache/
Ignored: analysis/ms15_mofa_sample_wm_new_meta_cache/
Ignored: analysis/ms15_mofa_sample_wm_superclean_cache/
Ignored: analysis/ms15_patients_cache/
Ignored: analysis/ms15_patients_gm_cache/
Ignored: analysis/ms15_patients_sample_level_cache/
Ignored: analysis/ms15_patients_w_ms_cache/
Ignored: analysis/supp06_sccaf_cache/
Ignored: analysis/supp07_superclean_check_cache/
Ignored: analysis/supp09_ancombc_cache/
Ignored: analysis/supp09_ancombc_mixed_cache/
Ignored: analysis/supp09_ancombc_superclean_cache/
Ignored: analysis/supp10_muscat_cache/
Ignored: analysis/supp10_muscat_ctrl_gm_vs_wm_cache/
Ignored: analysis/supp10_muscat_heatmaps_cache/
Ignored: analysis/supp10_muscat_olg_pc1_cache/
Ignored: analysis/supp10_muscat_olg_pc2_cache/
Ignored: analysis/supp10_muscat_olg_pc_cache/
Ignored: analysis/supp10_muscat_regression_cache/
Ignored: analysis/supp10_muscat_soup_cache/
Ignored: code/._ms10_muscat_fns_recover.R
Ignored: code/.recovery/
Ignored: code/jobs/._muscat_run09_2021-10-11.slurm
Ignored: code/muscat_plan.txt
Ignored: data/
Ignored: figures/
Ignored: output/
Ignored: tmp/
Untracked files:
Untracked: Rplots.pdf
Untracked: analysis/supp09_ancombc_superclean.Rmd
Untracked: code/ancom_v2.1.R
Untracked: code/dev_check_eqtls_against_groupings_2021-10-15.R
Untracked: code/dev_check_genotyping_20211007.R
Untracked: code/dev_check_mt_in_soup_2021-10-18.R
Untracked: code/dev_de_w_contamation_2021-10-25.R
Untracked: code/glmmtmb_code_for_julien_2021-10-13.R
Untracked: code/jobs/muscat_run09_2021-10-11.slurm
Untracked: code/jobs/muscat_run09_2021-10-12.slurm
Untracked: code/jobs/muscat_run09_2021-10-13.slurm
Untracked: code/jobs/muscat_run09_2021-10-14.slurm
Untracked: code/jobs/muscat_run10_2021-10-11.slurm
Untracked: code/jobs/muscat_run10_2021-10-12.slurm
Untracked: code/jobs/muscat_run10_2021-10-13.slurm
Untracked: code/jobs/muscat_run11_2021-10-11.slurm
Untracked: code/jobs/muscat_run11_2021-10-14.slurm
Untracked: code/jobs/muscat_run11_2021-10-21.slurm
Untracked: code/jobs/muscat_run12_2021-10-11.slurm
Untracked: code/jobs/muscat_run12_2021-10-14.slurm
Untracked: code/jobs/muscat_run12_2021-10-21.slurm
Untracked: code/jobs/muscat_run19_2021-10-14.slurm
Untracked: code/metadata_update_plan_20210928.txt
Untracked: code/ms10_muscat_fns_recover.R
Untracked: code/ms10_slurm_jobs.py
Untracked: code/muscat_slurm_template.slurm
Untracked: code/negbin_std.stan
Untracked: code/negbin_w_contam.stan
Untracked: code/negbin_w_contam_v2.stan
Unstaged changes:
Modified: analysis/fig_muscat.Rmd
Modified: analysis/ms07_soup.Rmd
Modified: analysis/ms09_ancombc_mixed.Rmd
Modified: analysis/ms14_lesions.Rmd
Modified: analysis/ms15_mofa_sample_wm_superclean.Rmd
Modified: analysis/supp07_superclean_check.Rmd
Modified: analysis/supp09_ancombc.Rmd
Deleted: code/dev_checking_hacked_pb_2021-07-16.R
Modified: code/fig_muscat.R
Modified: code/jobs/muscat_job_2021-10-04_fine_gm.slurm
Modified: code/jobs/muscat_job_2021-10-04_fine_wm.slurm
Modified: code/ms07_soup.R
Modified: code/ms10_muscat_fns.R
Modified: code/ms10_muscat_runs.R
Modified: code/ms14_lesions.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ms07_soup.Rmd) and HTML (docs/ms07_soup.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 13941a1 | Macnair | 2021-10-18 | Update soup and pseudobulk calcs with final metadata |
| html | 13941a1 | Macnair | 2021-10-18 | Update soup and pseudobulk calcs with final metadata |
| Rmd | 15ed138 | Macnair | 2021-10-04 | Update soup analysis with final metadata |
| html | 15ed138 | Macnair | 2021-10-04 | Update soup analysis with final metadata |
| Rmd | 5b8dcb7 | Macnair | 2021-06-03 | Updated soup & pseudobulk calcs with final type_fine |
| html | 5b8dcb7 | Macnair | 2021-06-03 | Updated soup & pseudobulk calcs with final type_fine |
| Rmd | 58205c2 | Macnair | 2021-05-21 | Update with random effects and markers |
| html | 9852840 | Macnair | 2021-05-12 | Adding docs to repo for first time - massive! |
| Rmd | eef8a1c | Macnair | 2021-04-29 | Minor tweaks to allow rerunning on Roche servers |
| Rmd | 0bd2043 | Macnair | 2021-04-19 | Improved soup coding |
| Rmd | 129c53d | Macnair | 2021-04-16 | Renamed a lot of things to add ms07_soup |
Setup / definitions
Libraries
Helper functions
source('code/ms00_utils.R')
source('code/ms03_SampleQC.R')
source('code/ms04_conos.R')
source('code/ms07_soup.R')Inputs
sce_f = "data/sce_raw/ms_sce.rds"
labels_f = "data/byhand_markers/validation_markers_2021-05-31.csv"
labelled_f = "output/ms13_labelling/conos_labelled_2021-05-31.txt.gz"
meta_f = "data/metadata/metadata_checked_assumptions_2021-10-08.xlsx"
gtf_f = "data/gtf/Homo_sapiens.GRCh38.96.filtered.preMRNA.gtf"
soup_f = "data/ambient/ambient.100UMI.txt"
qc_dir = "output/ms03_SampleQC"
qc_f = file.path(qc_dir, "ms_qc_dt.txt")Outputs
# set up directory
save_dir = 'output/ms07_soup'
date_tag = '2021-10-11'
if (!dir.exists(save_dir))
dir.create(save_dir)
# define markers for distinguishing broad celltypes
fm_broad_f = sprintf("%s/fm_broad_for_soup_%s_all_%s.txt", save_dir, "%s", date_tag)
pval_type = 'all'
tests = c('binom', 't', 'wilcox')
n_cells = 2000
n_cores = 8
# which to use?
top_rank = 10
sel_test = 'binom'
fdr_cut = 1e-6
logfc_cut = 2
# define pseudobulk files
exp_fine_f = sprintf('%s/pb_sum_fine_%s.rds', save_dir, date_tag)
det_fine_f = sprintf('%s/pb_prop_fine_%s.rds', save_dir, date_tag)
exp_broad_f = sprintf('%s/pb_sum_broad_%s.rds', save_dir, date_tag)
det_broad_f = sprintf('%s/pb_prop_broad_%s.rds', save_dir, date_tag)
# define soup files
soup_q_pat = sprintf('%s/soup_quantities_%s_%s_%s.rds', save_dir,
'%s', '%s', date_tag)
soup_broad_f = sprintf('%s/pb_soup_broad_%s_%s.rds', save_dir,
'maximum', date_tag)
soup_fine_f = sprintf('%s/pb_soup_fine_%s_%s.rds', save_dir,
'maximum', date_tag)
soup_broad_mito_f = sprintf('%s/pb_soup_broad_%s_%s.rds', save_dir,
'mito', date_tag)
soup_fine_mito_f = sprintf('%s/pb_soup_fine_%s_%s.rds', save_dir,
'mito', date_tag)Load inputs
conos_dt = load_labelled_dt(labelled_f, labels_f)
labels_dt = load_names_dt(labels_f)meta_dt = load_meta_dt_from_xls(meta_f, outlier_samples = NULL)
qc_stats = calc_qc_stats(qc_dir, qc_f, conos_dt)gtf_dt = load_gtf_dt(gtf_f)# do pseudobulking
pb_broad = make_pb_object(exp_broad_f, sce_f, meta_dt,
conos_dt[, .(cell_id, sample_id, type_broad)],
cluster_var = 'type_broad', fun = 'sum', n_cores = n_cores)loading pre-saved objectprops_broad = make_pb_object(det_broad_f, sce_f, meta_dt,
conos_dt[, .(cell_id, sample_id, type_broad)],
cluster_var = 'type_broad', fun = 'prop.detected', n_cores = n_cores)loading pre-saved objectpb_fine = make_pb_object(exp_fine_f, sce_f, meta_dt,
conos_dt[, .(cell_id, sample_id, type_fine)],
cluster_var = 'type_fine', fun = 'sum', n_cores = n_cores)loading pre-saved objectprops_fine = make_pb_object(det_fine_f, sce_f, meta_dt,
conos_dt[, .(cell_id, sample_id, type_fine)],
cluster_var = 'type_fine', fun = 'prop.detected', n_cores = n_cores)loading pre-saved objectsoup_dt = soup_f %>% fread %>%
.[, gene_id := paste0(symbol, '_', ensembl)] %>% setcolorder('gene_id')
assert_that(all(rownames(pb_fine) == soup_dt$gene_id))[1] TRUEassert_that(all(colnames(pb_fine) %in% names(soup_dt)[-seq.int(3)]))[1] TRUEProcessing / calculations
Calc find markers for broad celltypes
set.seed(20211011)
fm_fs = sapply(tests, function(t) sprintf(fm_broad_f, t))
calc_find_markers_soup(sce_f, fm_fs, tests, pval_type, conos_dt, n_cells, n_cores)already done! skippingNULL# define list to keep
exc_str = '(pseudogene|antisense|lincRNA)'
proper_gs = gtf_dt[!str_detect(gene_biotype, exc_str)]$symbol
top_fms = lapply(tests,
function(t) find_top_markers(fm_broad_f, t, proper_gs, top_rank)) %>%
setNames(tests)
# define list for
marker_list = calc_broad_marker_list(fm_broad_f, sel_test, proper_gs,
fdr_cut = fdr_cut, val_cut = logfc_cut)Estimate contamination
prof_fine = calc_soup_profile(pb_fine, soup_dt)
prof_broad = calc_soup_profile(pb_broad, soup_dt)# make list of objects
pb_list = list(type_fine = pb_fine, type_broad = pb_broad)
prof_list = list(prof_fine, prof_broad) %>% setNames(names(pb_list))
for (t in names(pb_list)) {
# which pseudobulk object?
this_pb = pb_list[[t]]
this_prof = prof_list[[t]]
# estimate maximum soup
soup_q_f = sprintf(soup_q_pat, t, 'maximum')
soup_max = calc_soup_quantities(soup_q_f, this_pb, this_prof,
soup_method = 'maximum', n_cores = n_cores, n_iters = 5, n_points = 10)
# estimate soup by broad markeers
soup_q_f = sprintf(soup_q_pat, t, 'control')
soup_ctrl = calc_soup_quantities(soup_q_f, this_pb, this_prof,
soup_method = 'control', marker_list = marker_list, n_cores = n_cores)
# estimate mito soup
soup_q_f = sprintf(soup_q_pat, t, 'mito')
soup_mt = calc_soup_quantities(soup_q_f, this_pb, this_prof,
soup_method = 'mito', n_cores = n_cores, n_iters = 5, n_points = 10)
}already done!
already done!
already done!
already done!
already done!
already done!# mito soup calculations
soup_mt = sprintf(soup_q_pat, 'type_broad', 'mito') %>% readRDS
pb_soup = calc_pb_soup(pb_broad, soup_mt, prof_broad)
mean_mt = calc_contaminated_genes(pb_soup)
# print some outputs
message('total non-contaminated genes by broad celltype, mito calc:')total non-contaminated genes by broad celltype, mito calc:colSums(mean_mt <= 0.1, na.rm = TRUE) %>% sort %>% round(2) %>% printExcitatory neurons Inhibitory neurons Oligodendrocytes Microglia
113 1396 1643 7276
Endothelial cells Astrocytes Immune Pericytes
8111 9036 10464 11290
OPCs / COPs
16222 message('proportion non-contaminated genes by broad celltype, mito calc:')proportion non-contaminated genes by broad celltype, mito calc:colMeans(mean_mt <= 0.1, na.rm = TRUE) %>% sort %>% round(2) %>% printExcitatory neurons Inhibitory neurons Oligodendrocytes Microglia
0.00 0.05 0.05 0.25
Endothelial cells Astrocytes Immune Pericytes
0.29 0.30 0.41 0.43
OPCs / COPs
0.56 # "maximum" soup calculations
soup_max = sprintf(soup_q_pat, 'type_broad', 'maximum') %>% readRDS
pb_soup = calc_pb_soup(pb_broad, soup_max, prof_broad)
mean_cont = calc_contaminated_genes(pb_soup)
# print some outputs
message('total non-contaminated genes by broad celltype:')total non-contaminated genes by broad celltype:colSums(mean_cont <= 0.1, na.rm = TRUE) %>% sort %>% round(2) %>% print Immune Pericytes Microglia Endothelial cells
17349 19561 21843 22469
OPCs / COPs Oligodendrocytes Astrocytes Inhibitory neurons
23424 24556 25161 26931
Excitatory neurons
28811 message('proportion non-contaminated genes by broad celltype:')proportion non-contaminated genes by broad celltype:colMeans(mean_cont <= 0.1, na.rm = TRUE) %>% sort %>% round(2) %>% print Immune Microglia Pericytes Endothelial cells
0.68 0.74 0.74 0.79
Oligodendrocytes OPCs / COPs Astrocytes Inhibitory neurons
0.80 0.81 0.83 0.89
Excitatory neurons
0.92 # mito soup calculations
soup_mt = sprintf(soup_q_pat, 'type_fine', 'mito') %>% readRDS
pb_soup = calc_pb_soup(pb_fine, soup_mt, prof_fine)
mean_mt = calc_contaminated_genes(pb_soup)
# print some outputs
message('total non-contaminated genes by fine celltype, mito calc:')total non-contaminated genes by fine celltype, mito calc:colSums(mean_mt <= 0.1, na.rm = TRUE) %>% sort %>% round(2) %>% print Ex_RORB_CUX2_A Oligo_A2 Ex_RORB_A Ex_CUX2_B
1620 1700 1813 2692
Ex_RORB_CUX2_B Inh_Pvalb_A Ex_THEMIS_A Neuro_oligo
3449 3917 4005 4290
Ex_TLE4_A Ex_RORB_CUX2_C Ex_RORB_CUX2_D Microglia_B
4667 5443 5568 5709
Ex_RORB_THEMIS_B Ex_THEMIS_B Oligo_B3 Microglia_A
5755 6546 7390 7640
Ex_RORB_B Ex_RORB_CUX2_F Ex_RORB_THEMIS_A Astro_A
7707 8073 8097 8423
Endo_B Oligo_B4 Ex_CUX2_A Inh_Pvalb_B
8484 9050 9068 9070
Oligo_D Inh_SST_B Inh_SST_A Ex_TLE4_B
9147 9248 9551 9568
Endo_A T_cells Inh_VIP Ex_RORB_C
9734 10044 10180 10208
Oligo_C1 Oligo_A1 Astro_C Inh_LAMP5
10236 10241 10247 10291
Astro_D B_cells Ex_RORB_CUX2_E Inh_RELN
10445 10632 10715 10734
Astro_E Pericytes COP_A1 Oligo_B1
11190 11290 12363 12475
COP_B Oligo_B2 Oligo_C2 COP_A2
13261 14858 15659 16353
Astro_B OPC
16871 19874 message('proportion non-contaminated genes by fine celltype, mito calc:')proportion non-contaminated genes by fine celltype, mito calc:colMeans(mean_mt <= 0.1, na.rm = TRUE) %>% sort %>% round(2) %>% print Ex_RORB_CUX2_A Oligo_A2 Ex_RORB_A Ex_CUX2_B
0.06 0.06 0.06 0.09
Ex_RORB_CUX2_B Inh_Pvalb_A Ex_THEMIS_A Ex_TLE4_A
0.12 0.14 0.14 0.16
Neuro_oligo Ex_RORB_CUX2_C Ex_RORB_CUX2_D Ex_RORB_THEMIS_B
0.17 0.19 0.20 0.20
Ex_THEMIS_B Oligo_B3 Microglia_A Ex_RORB_B
0.24 0.26 0.26 0.27
Microglia_B Astro_A Ex_RORB_THEMIS_A Ex_RORB_CUX2_F
0.27 0.28 0.29 0.30
Oligo_B4 Ex_CUX2_A Endo_B Oligo_D
0.31 0.31 0.32 0.33
Inh_SST_B Inh_SST_A Oligo_C1 Inh_VIP
0.34 0.35 0.35 0.35
Endo_A Ex_TLE4_B Inh_Pvalb_B Ex_RORB_C
0.35 0.36 0.36 0.37
Inh_LAMP5 Astro_C Oligo_A1 Inh_RELN
0.37 0.38 0.39 0.39
Ex_RORB_CUX2_E Astro_D T_cells Pericytes
0.40 0.40 0.42 0.43
Oligo_B1 COP_A1 B_cells Oligo_B2
0.45 0.47 0.48 0.52
Astro_E COP_B Astro_B Oligo_C2
0.52 0.57 0.59 0.69
OPC COP_A2
0.69 0.74 # use max value
soup_max = sprintf(soup_q_pat, 'type_fine', 'maximum') %>% readRDS
pb_soup = calc_pb_soup(pb_fine, soup_max, prof_fine)
mean_cont = calc_contaminated_genes(pb_soup)
# print some outputs
message('total non-contaminated genes by fine celltype:')total non-contaminated genes by fine celltype:colSums(mean_cont <= 0.1, na.rm = TRUE) %>% sort %>% round(2) %>% print Neuro_oligo Microglia_B Oligo_A1 Astro_E
10883 11315 13194 13368
COP_B B_cells COP_A1 Ex_RORB_CUX2_E
14624 14710 15873 15993
COP_A2 Astro_C Oligo_C2 T_cells
16074 16090 16200 16275
Inh_Pvalb_B Ex_RORB_CUX2_F Oligo_D Inh_SST_B
16281 16509 18596 19005
Oligo_A2 Endo_A Inh_SST_A Pericytes
19368 19393 19436 19561
Ex_THEMIS_B Oligo_B1 Inh_RELN Ex_RORB_CUX2_A
20383 20527 21211 21292
Ex_TLE4_B Inh_LAMP5 Microglia_A Ex_RORB_THEMIS_A
21654 21859 21861 21990
Astro_D Oligo_B2 Ex_RORB_C Oligo_B3
22213 22426 22529 22591
Endo_B Ex_RORB_CUX2_D Inh_Pvalb_A Astro_A
22633 23347 23428 23654
Ex_TLE4_A Oligo_B4 Inh_VIP Ex_RORB_THEMIS_B
23786 23829 23907 24099
Ex_RORB_B Astro_B Oligo_C1 Ex_THEMIS_A
24425 24517 24562 24738
OPC Ex_RORB_CUX2_B Ex_CUX2_A Ex_RORB_A
24774 25951 25995 26291
Ex_RORB_CUX2_C Ex_CUX2_B
26354 27222 message('proportion non-contaminated genes by fine celltype:')proportion non-contaminated genes by fine celltype:colMeans(mean_cont <= 0.1, na.rm = TRUE) %>% sort %>% round(2) %>% print Neuro_oligo Oligo_A1 Microglia_B Astro_C
0.44 0.50 0.54 0.60
COP_A1 Ex_RORB_CUX2_E Ex_RORB_CUX2_F Astro_E
0.60 0.60 0.62 0.63
COP_B Inh_Pvalb_B Oligo_D B_cells
0.63 0.65 0.66 0.67
Oligo_A2 T_cells Inh_SST_B Inh_SST_A
0.67 0.67 0.69 0.70
Endo_A Oligo_C2 COP_A2 Ex_RORB_CUX2_A
0.71 0.71 0.73 0.73
Oligo_B1 Ex_THEMIS_B Pericytes Microglia_A
0.74 0.74 0.74 0.74
Inh_RELN Oligo_B2 Astro_A Inh_LAMP5
0.77 0.78 0.79 0.79
Oligo_B3 Ex_RORB_THEMIS_A Ex_RORB_C Ex_TLE4_B
0.79 0.79 0.81 0.81
Oligo_B4 Ex_TLE4_A Inh_Pvalb_A Inh_VIP
0.81 0.82 0.82 0.83
Ex_RORB_CUX2_D Endo_B Oligo_C1 Astro_B
0.84 0.84 0.85 0.85
Ex_RORB_B Ex_THEMIS_A Ex_RORB_THEMIS_B Astro_D
0.85 0.85 0.86 0.86
OPC Ex_CUX2_A Ex_RORB_A Ex_RORB_CUX2_B
0.86 0.89 0.89 0.89
Ex_RORB_CUX2_C Ex_CUX2_B
0.90 0.91 # estimate maximum soup
soup_q_f = sprintf(soup_q_pat, 'type_fine', 'max_random')
set.seed(20211011)
mt_idx = rownames(prof_fine) %>% str_detect('^(MT-|MALAT1)')
assert_that(sum(mt_idx) == 14)[1] TRUEprof_rand = prof_fine %>% apply(2, function(col) {
# make output
out = numeric(length(col))
# fix the mito reads
out[mt_idx] = col[mt_idx]
# permute the rest
out[!mt_idx] = col[!mt_idx] %>% sample(length(.))
return(out)
}) %>% set_rownames(rownames(prof_fine))
soup_rand = calc_soup_quantities(soup_q_f, pb_fine, prof_rand,
soup_method = 'maximum', n_cores = n_cores, n_iters = 5, n_points = 10)already done!# get soup
soup_ctrl = sprintf(soup_q_pat, 'type_fine', 'control') %>% readRDS
soup_max = sprintf(soup_q_pat, 'type_fine', 'maximum') %>% readRDS
soup_mt = sprintf(soup_q_pat, 'type_fine', 'mito') %>% readRDS
soup_rand = sprintf(soup_q_pat, 'type_fine', 'max_random') %>% readRDS
# calc props
props_ctrl = calc_soup_props(pb_fine, soup_ctrl, meta_dt, labels_dt)
props_max = calc_soup_props(pb_fine, soup_max, meta_dt, labels_dt)
props_mt = calc_soup_props(pb_fine, soup_mt, meta_dt, labels_dt)
props_rand = calc_soup_props(pb_fine, soup_rand, meta_dt, labels_dt)Analysis
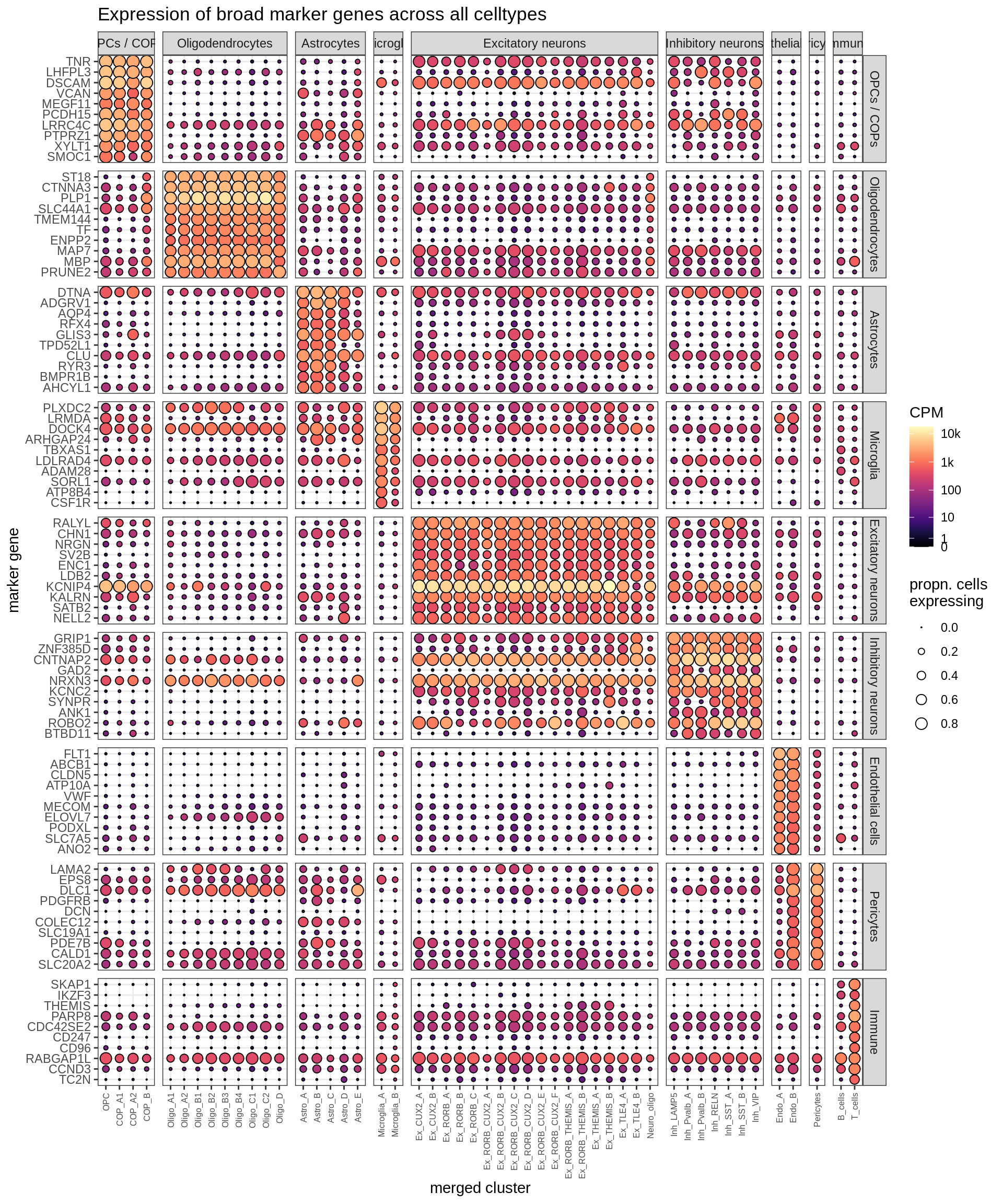
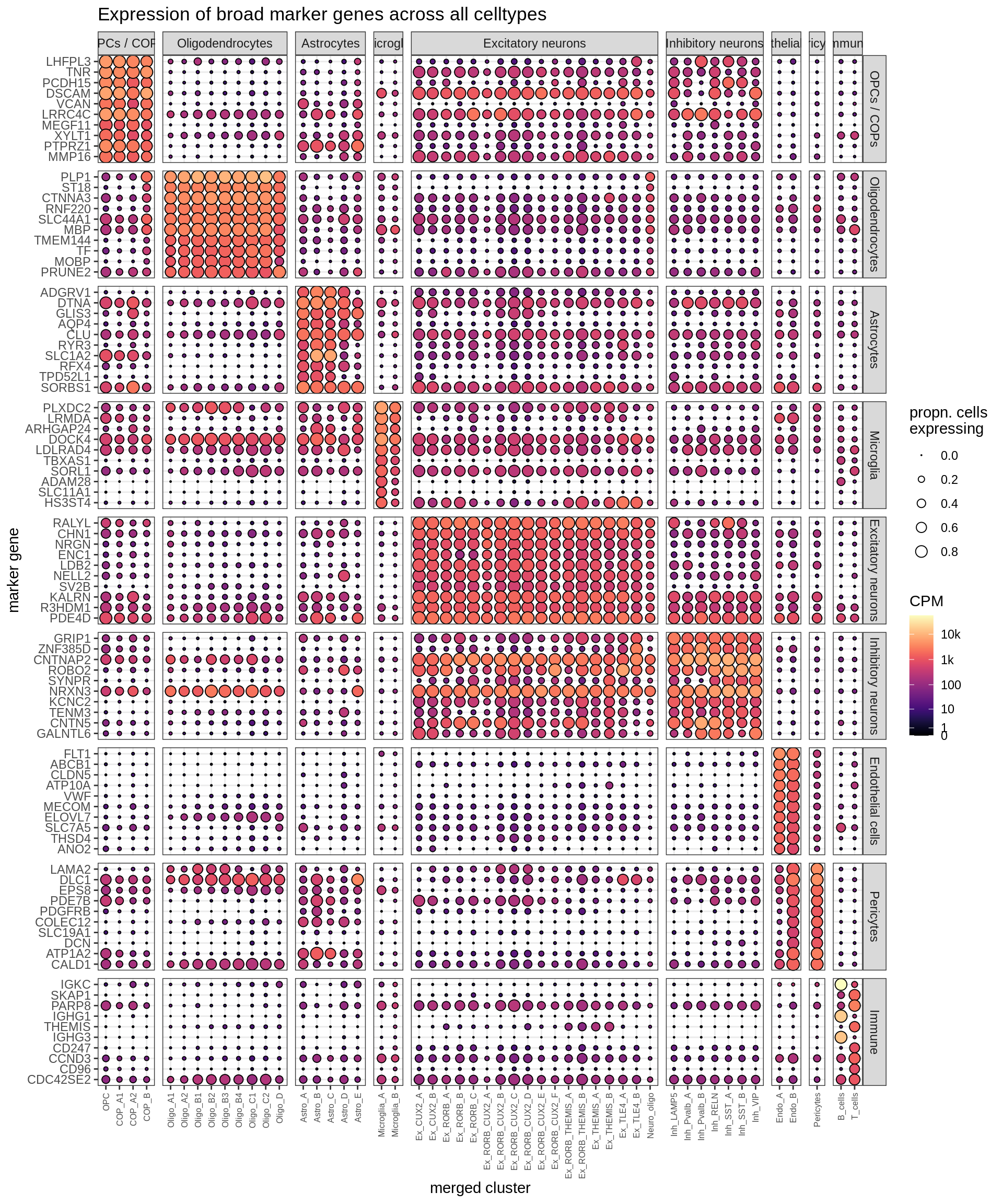
Dotplots of (calced) marker genes
for (t in tests) {
cat('### Markers via', t, 'test\n')
top_dt = top_fms[[t]]
suppressWarnings(print(plot_dotplot(pb_fine, props_fine, top_dt, labels_dt,
row_split = 'broad_marker')))
cat('\n\n')
}Markers via binom test
Markers via t test
How similar are soup estimates?
plot_soup_max_vs_ctrl(soup_max, soup_ctrl, labels_dt)Warning in sqrt(x): NaNs producedWarning: Removed 1603 rows containing missing values (geom_point).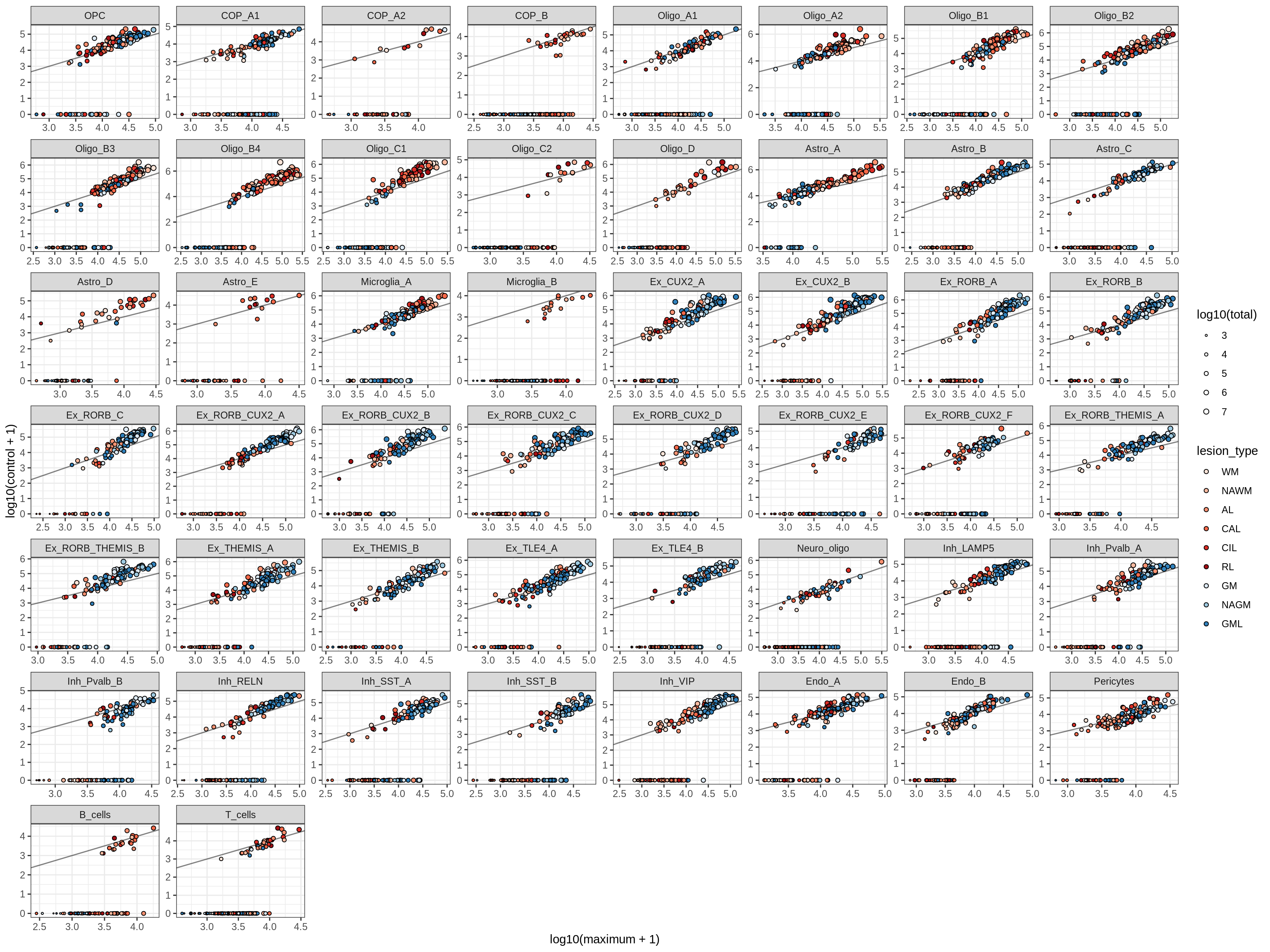
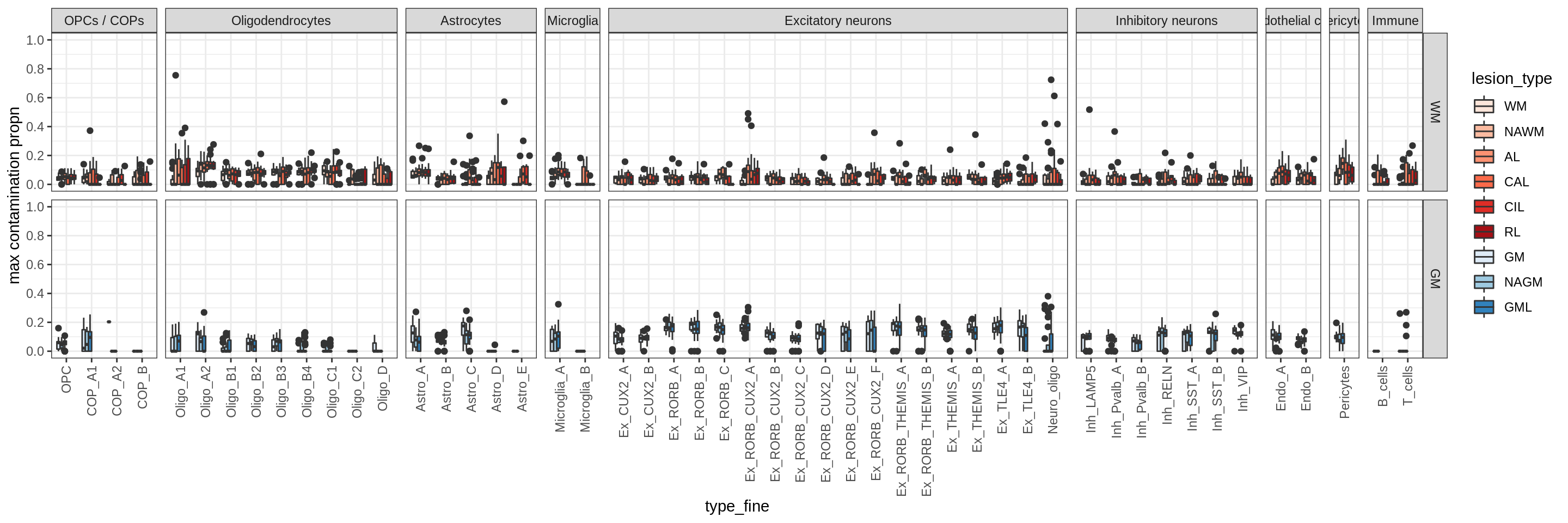
Distributions of soup proportions by cluster
cat('\n### Maximum soup')Maximum soup
print(plot_soup_contribution_by_cluster(props_max))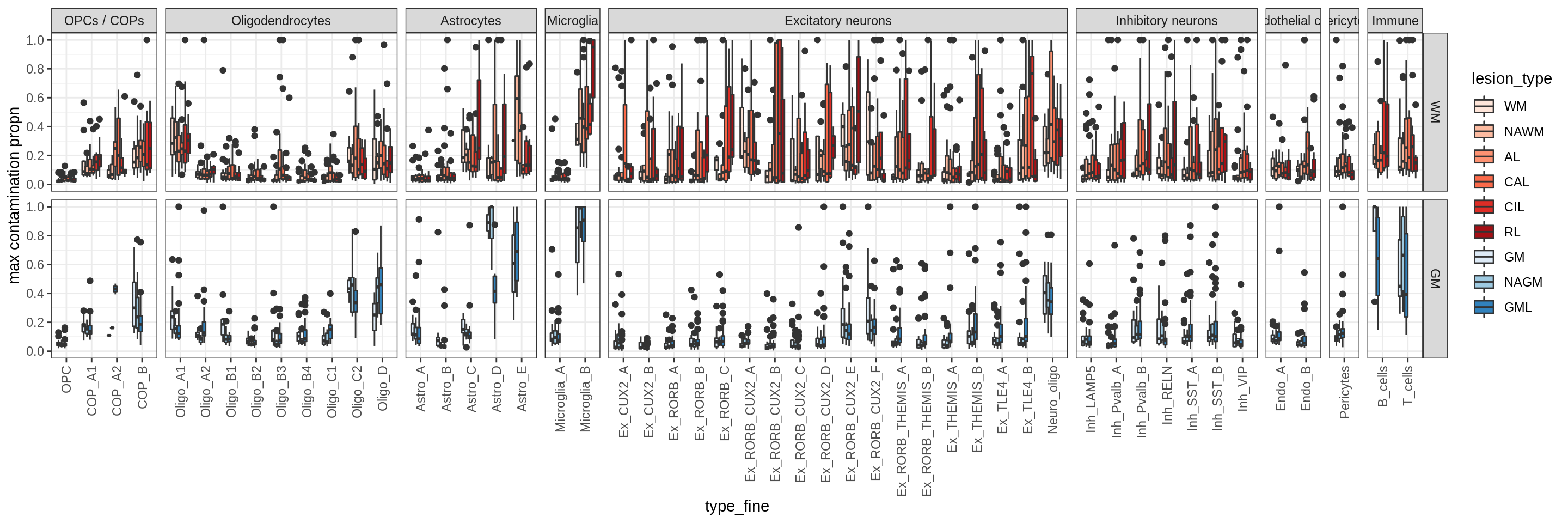
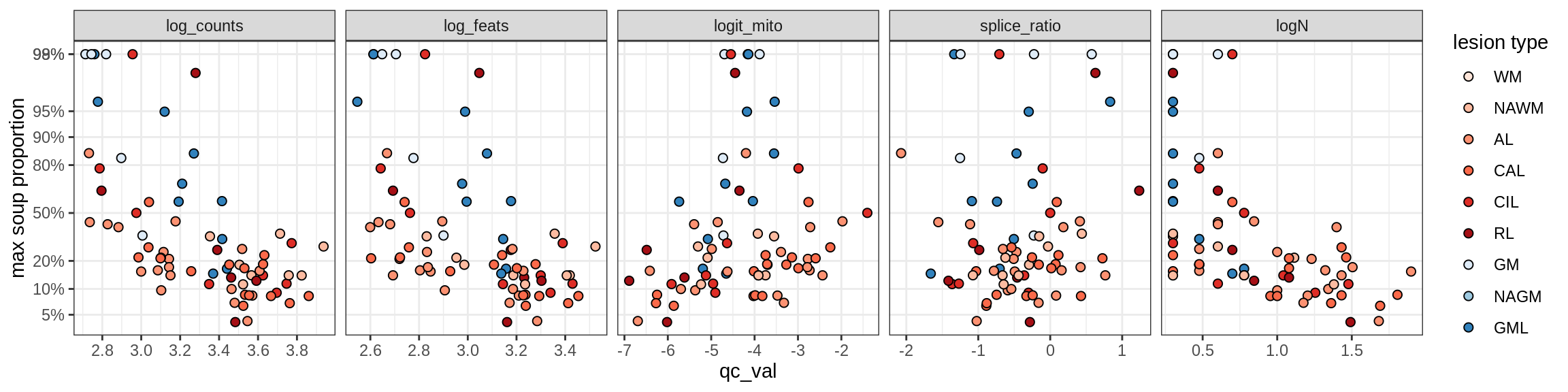
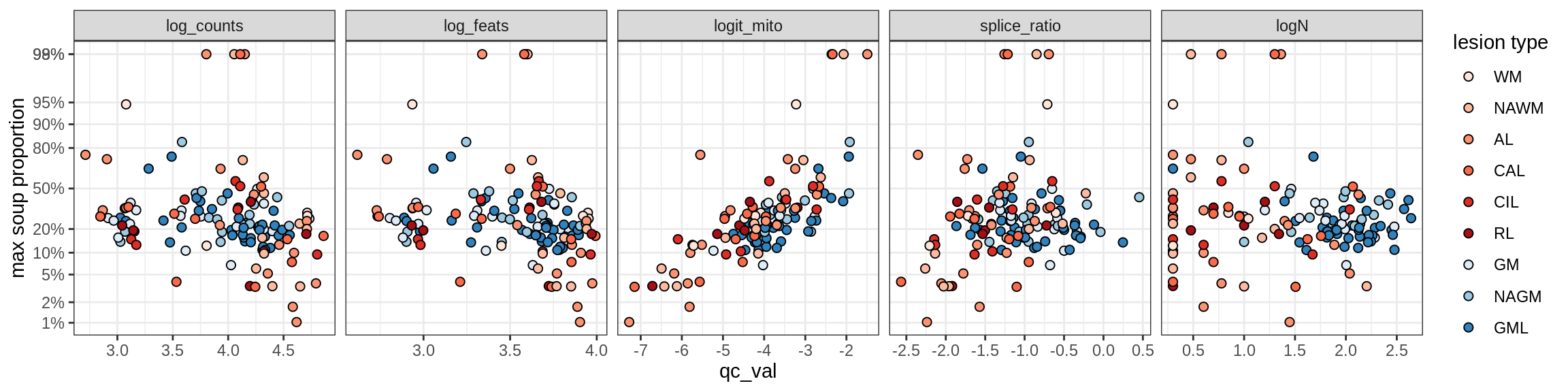
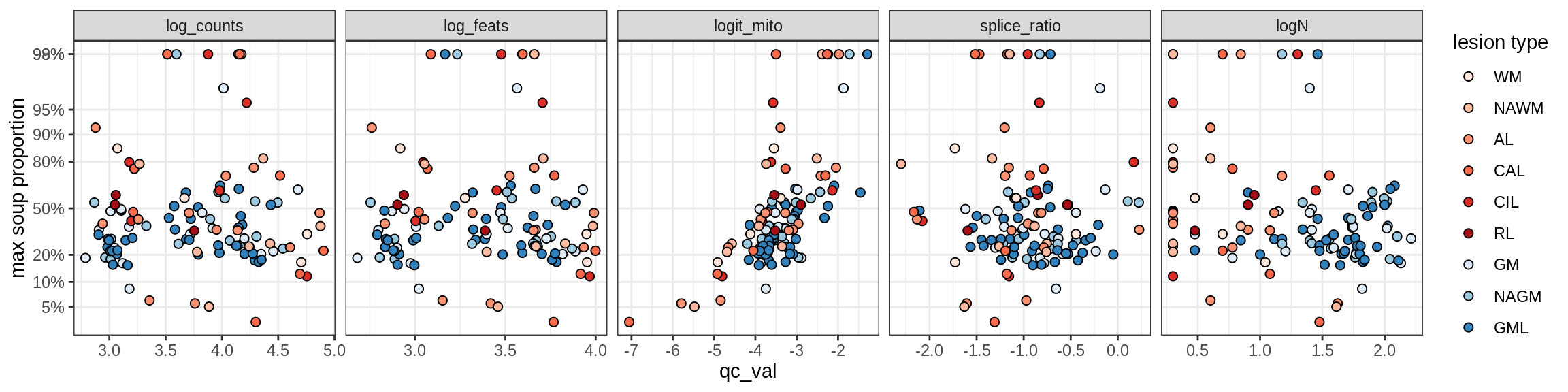
cat('\n\n')cat('\n### Celltype-based soup')Check of (celltype) contamination against QC metrics (oligos only)
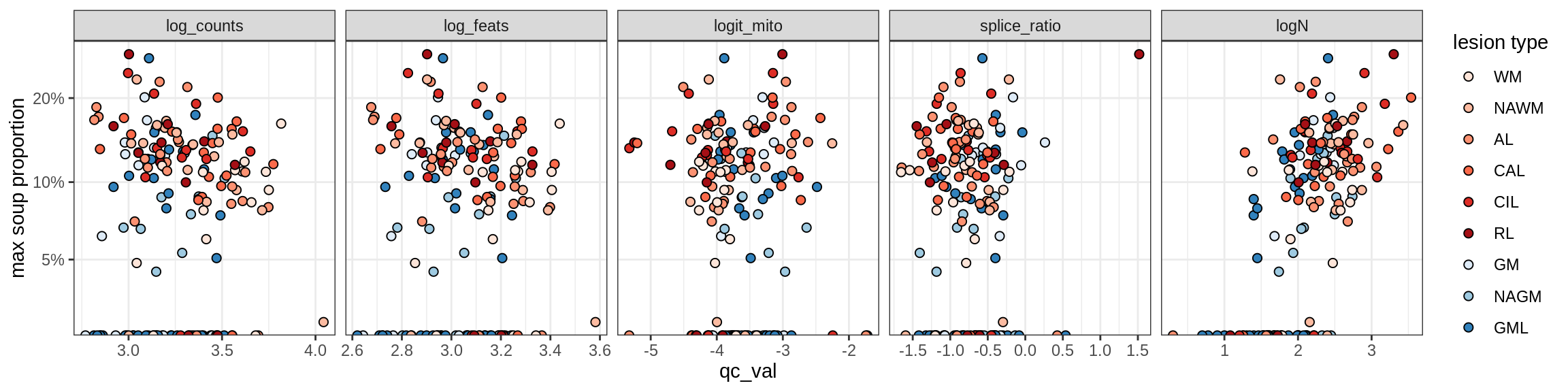
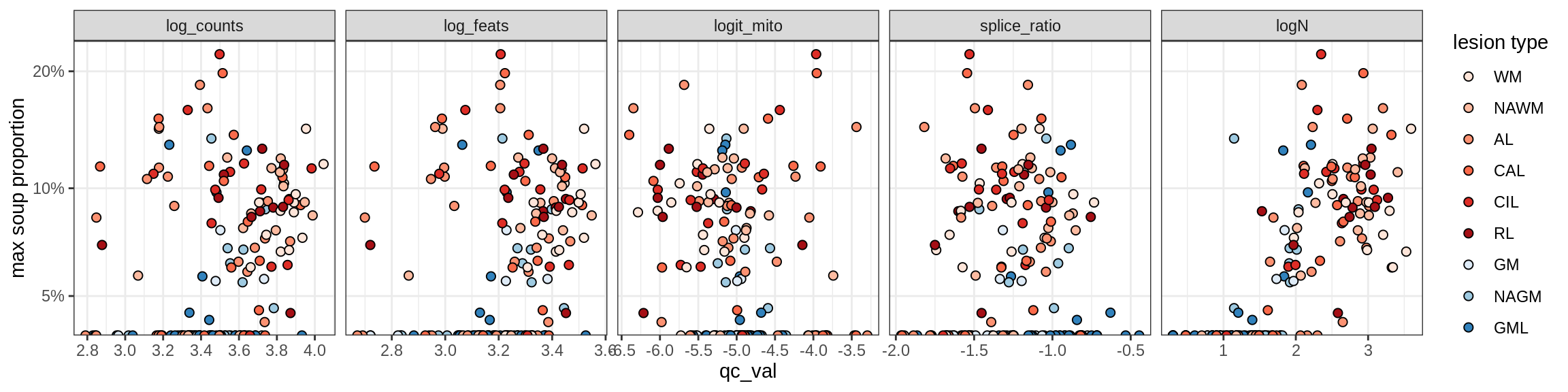
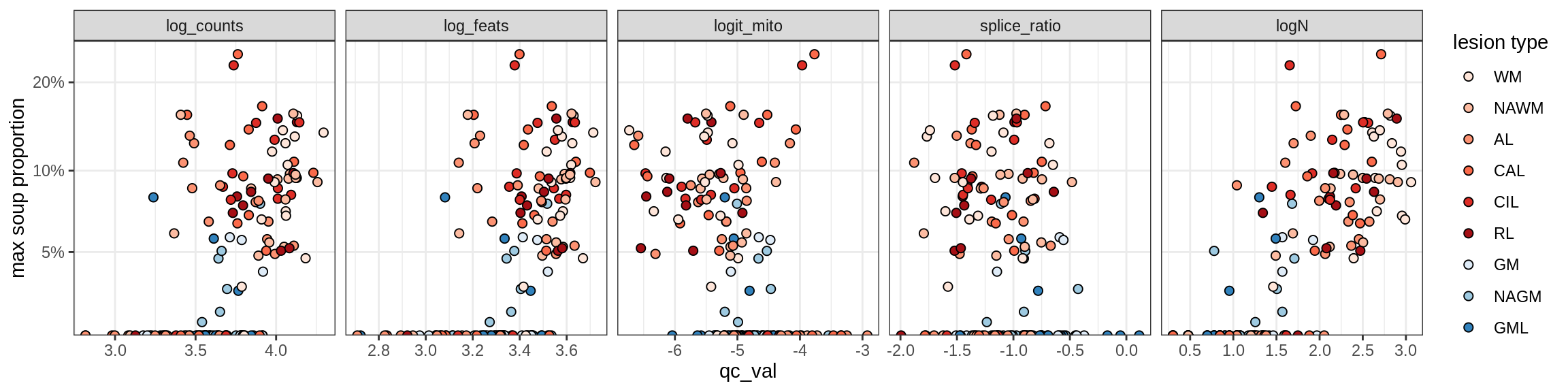
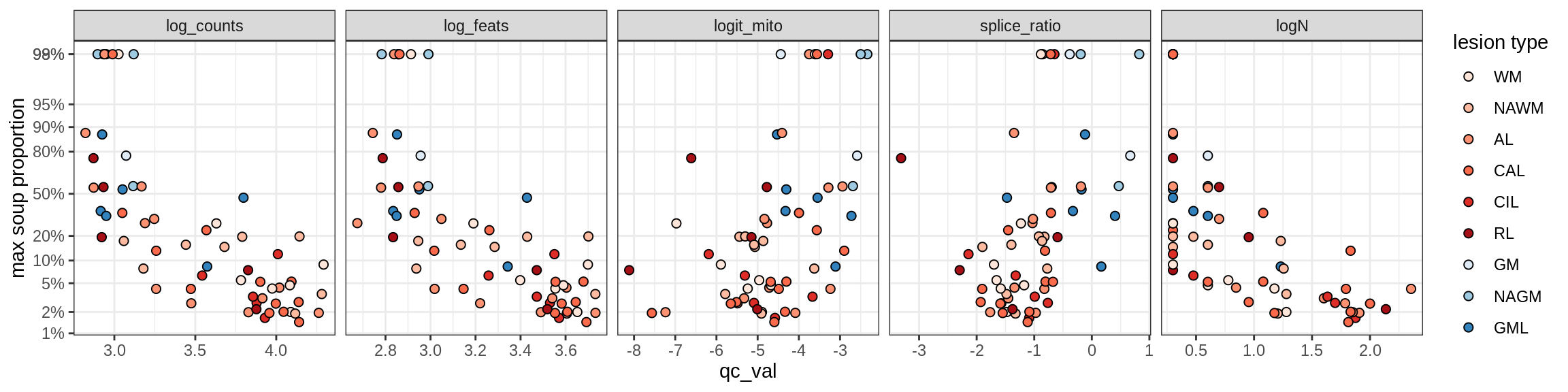
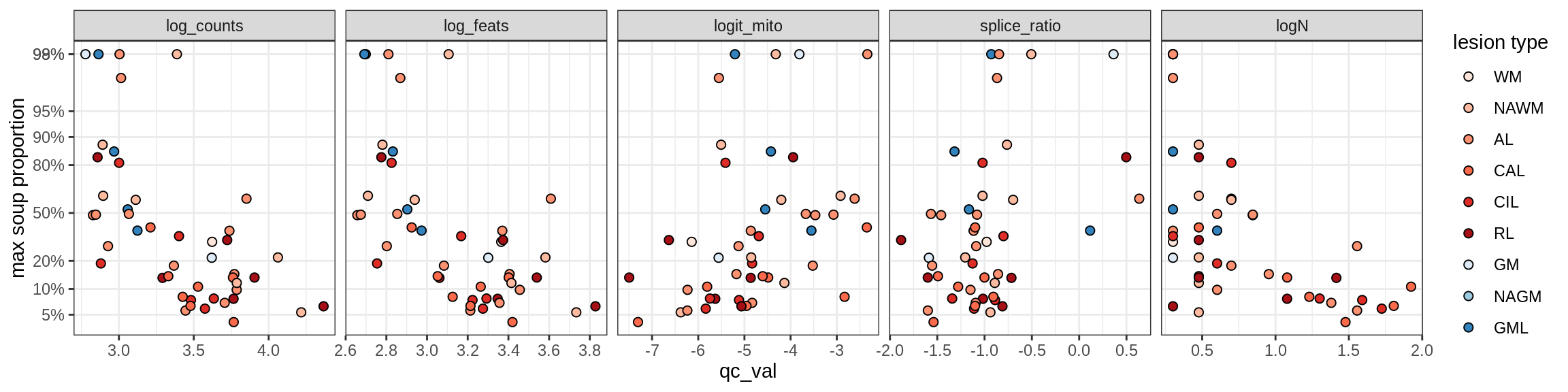
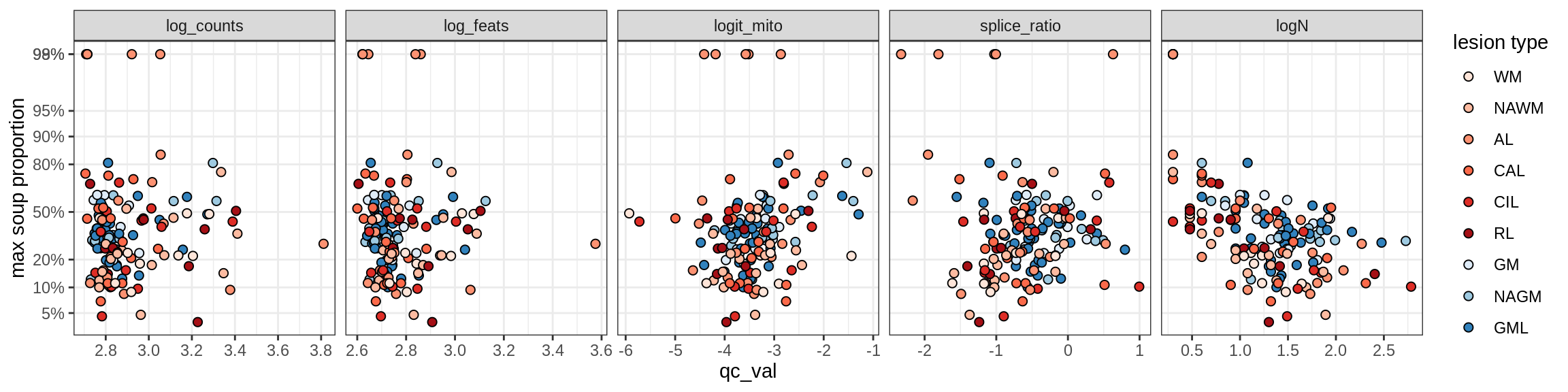
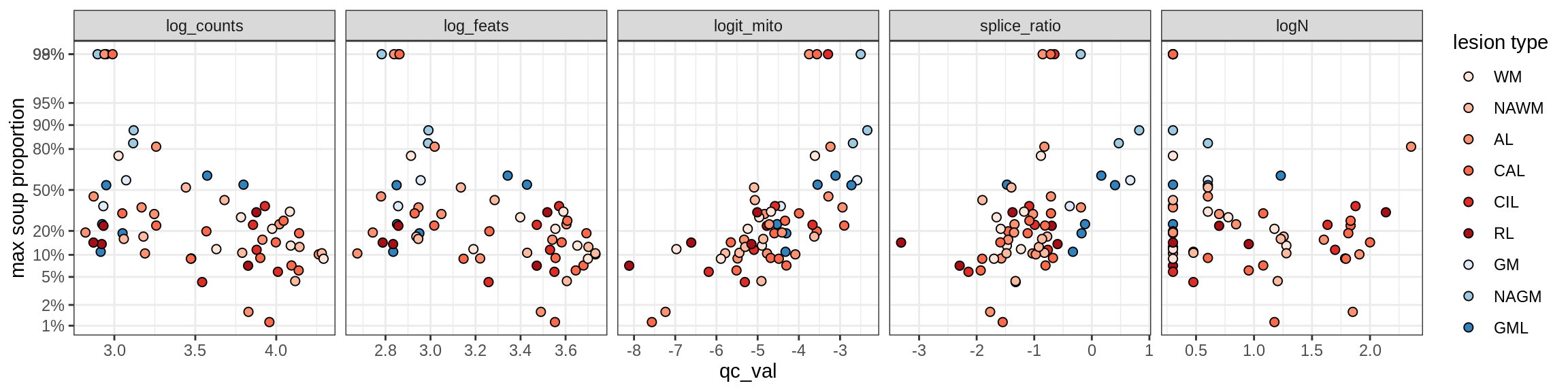
b = 'Oligodendrocytes'
for (t in unique(props_max[type_broad == b]$type_fine)) {
cat('### ', t, '\n')
suppressWarnings(print(plot_soup_props_vs_qc(props_ctrl[type_fine == t], qc_stats)))
cat('\n\n')
}Oligo_A1
Oligo_A2
Oligo_B1
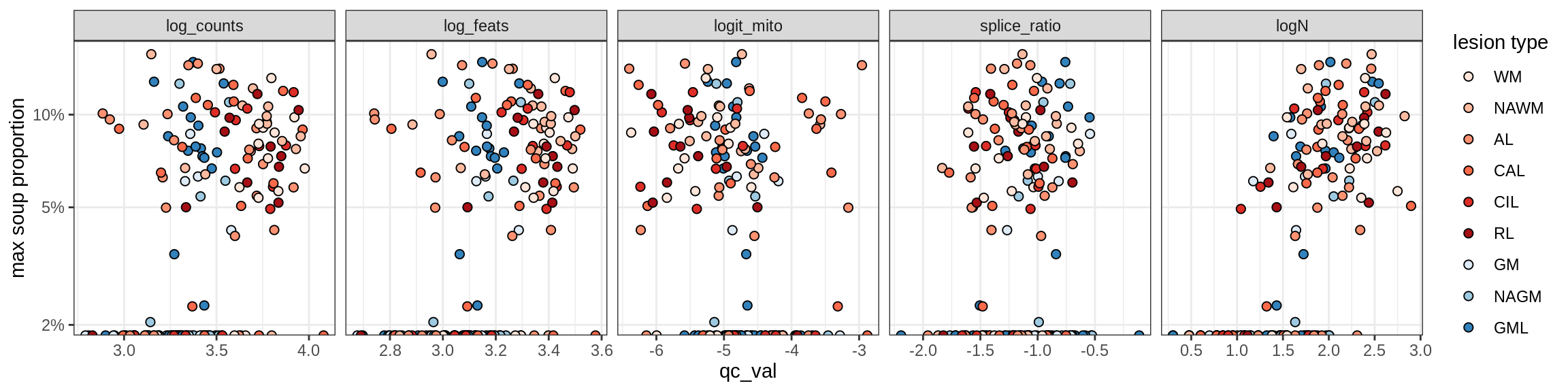
Oligo_B2
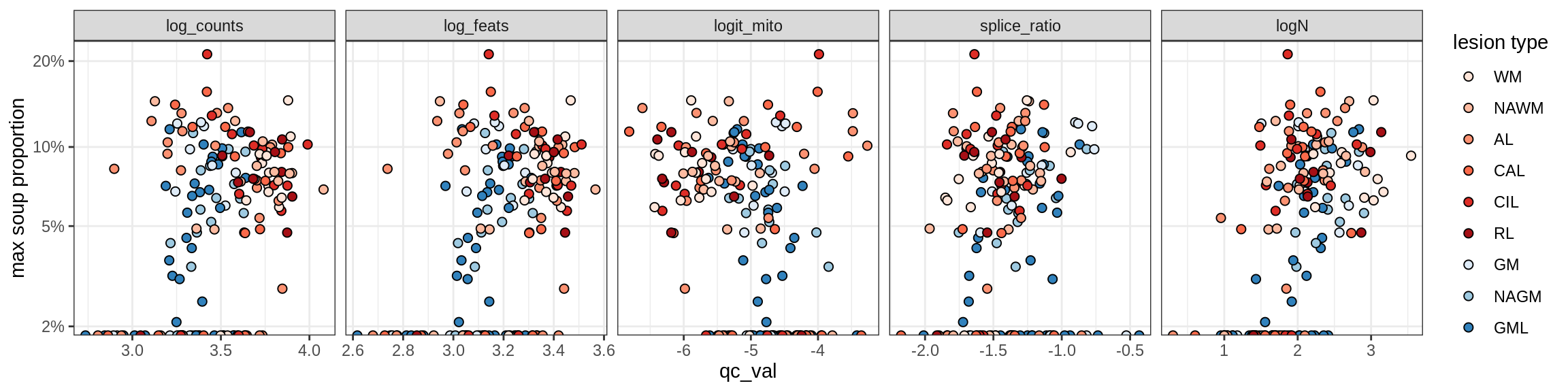
Oligo_B3
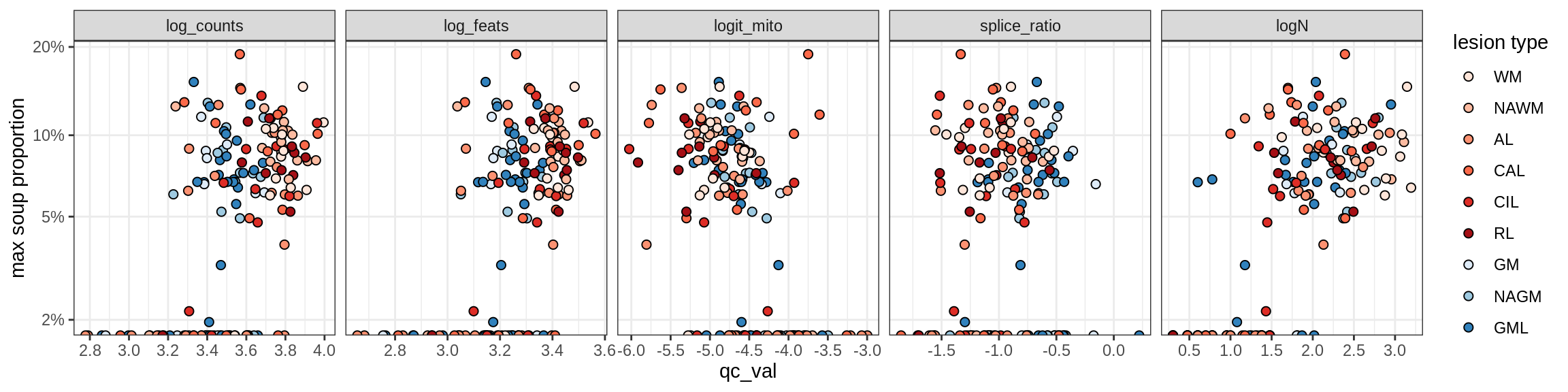
Oligo_B4
Oligo_C1
Oligo_C2
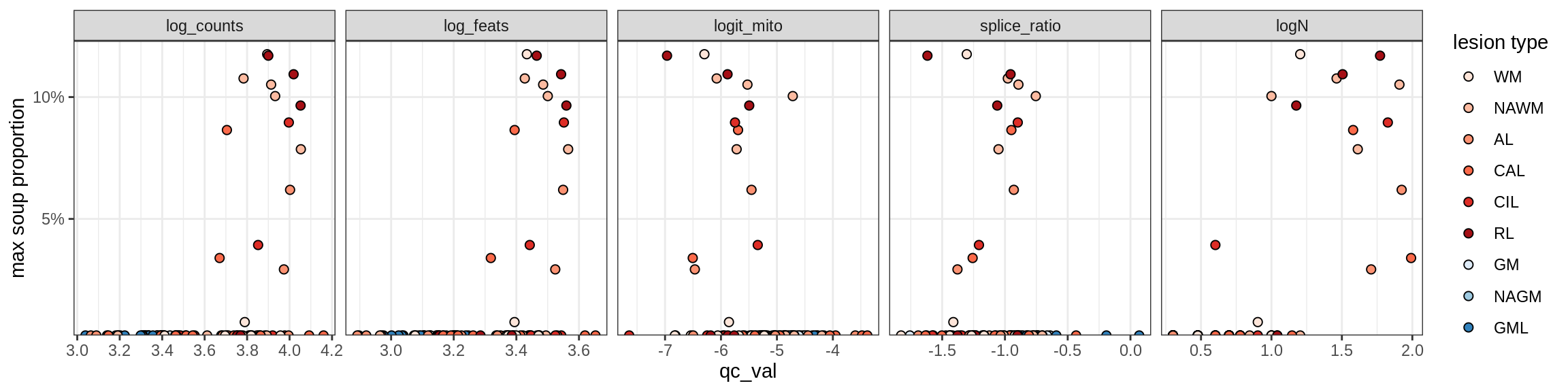
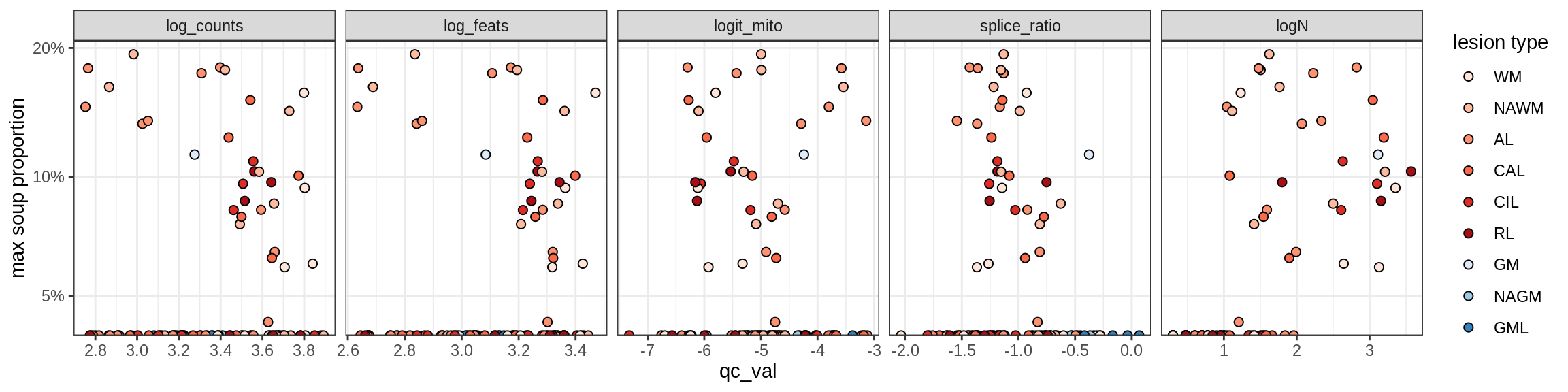
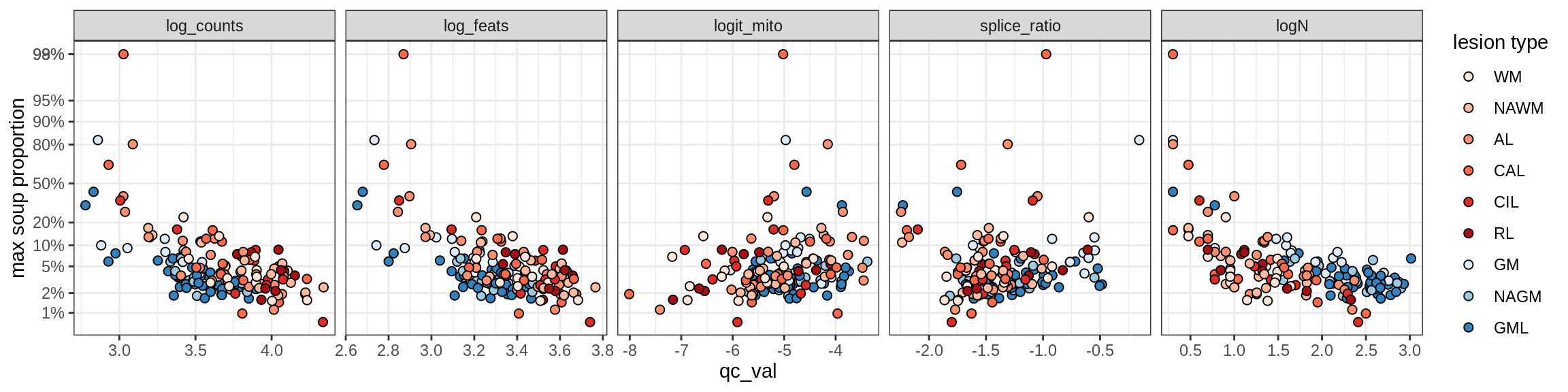
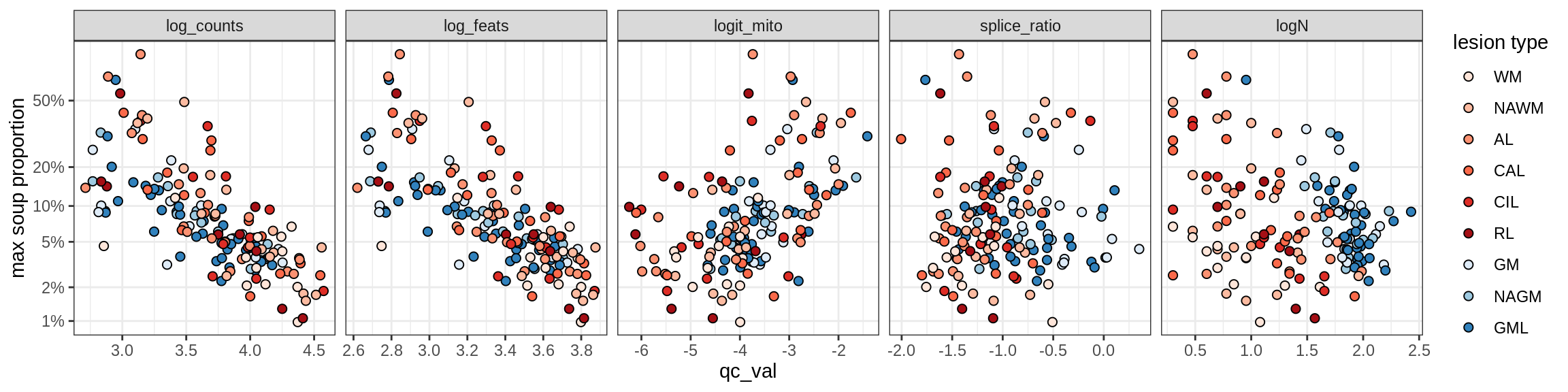
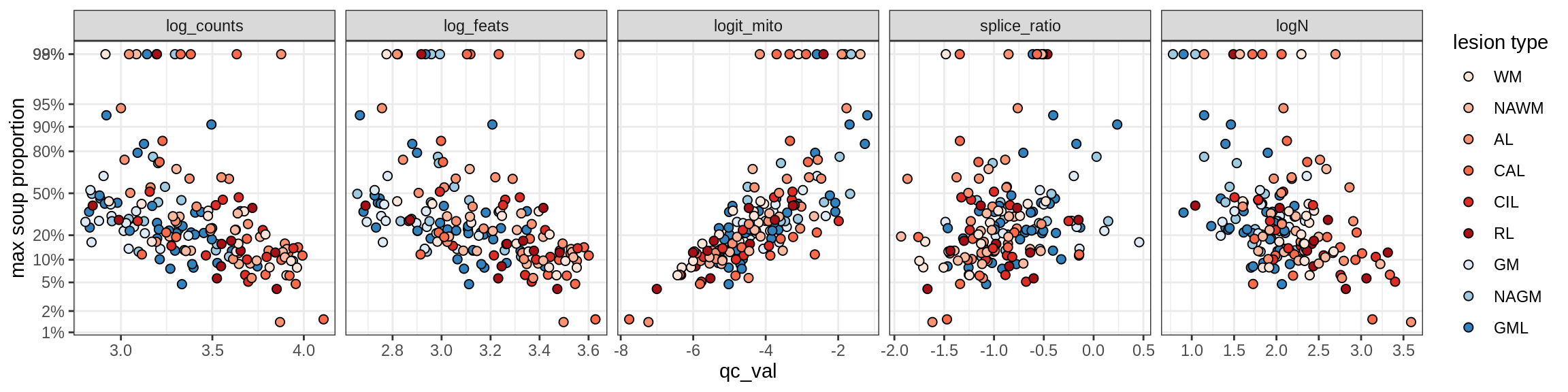
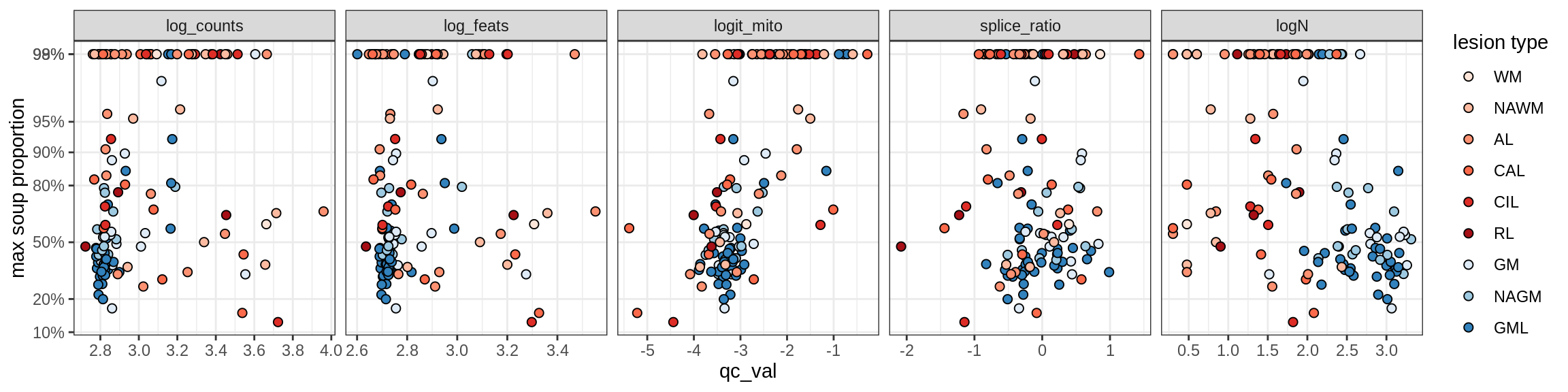
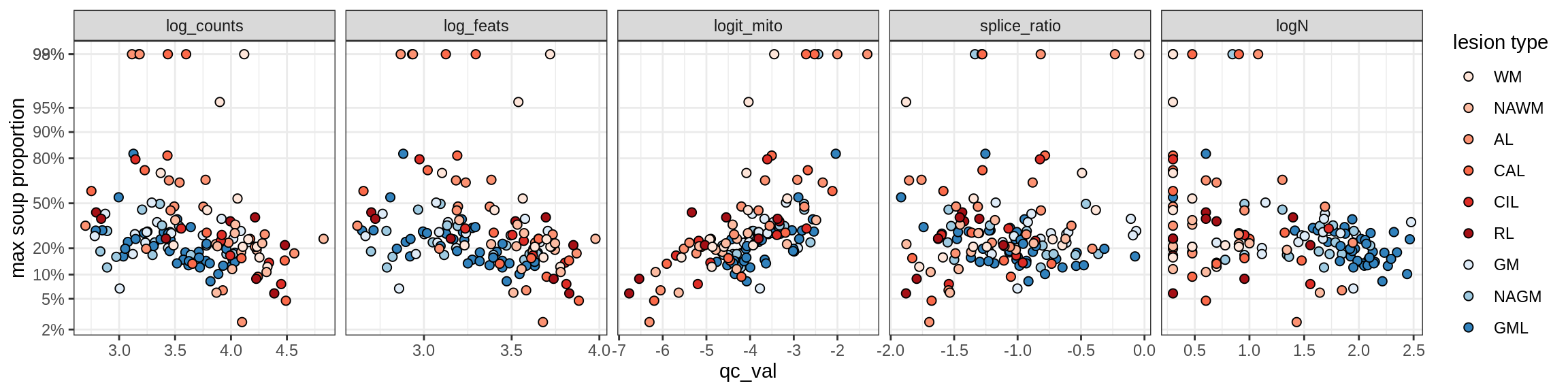
Correlations of “maximum” contamination with QC metrics
for (b in broad_ord) {
cat('### ', b, '{.tabset}\n')
for (t in levels(fct_drop(props_max[type_broad == b]$type_fine))) {
cat('#### ', t, '\n')
suppressWarnings(print(plot_soup_props_vs_qc(props_max[type_fine == t], qc_stats)))
cat('\n\n')
}
}OPCs / COPs
OPC
COP_A1
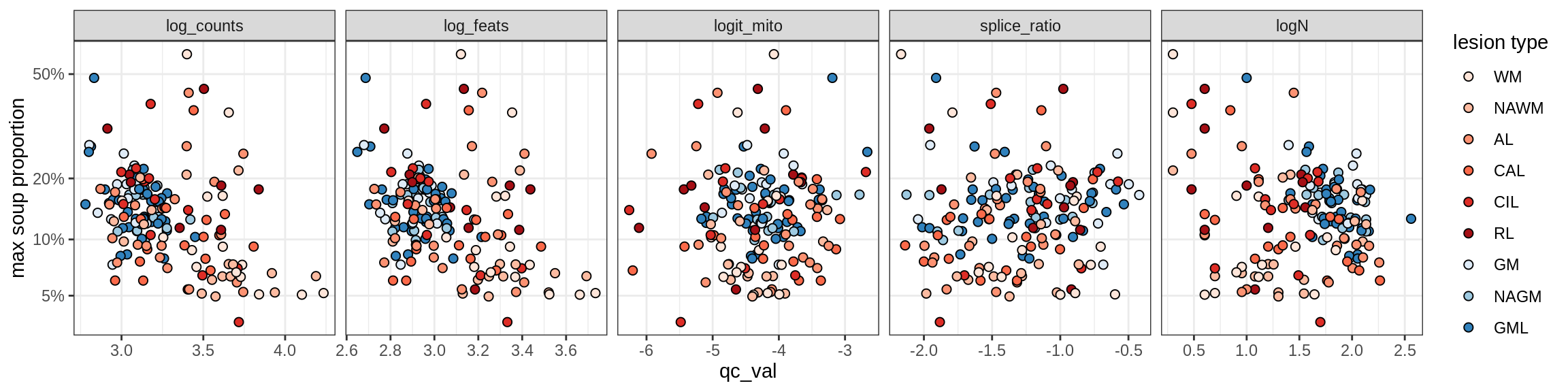
COP_A2
COP_B
Oligodendrocytes
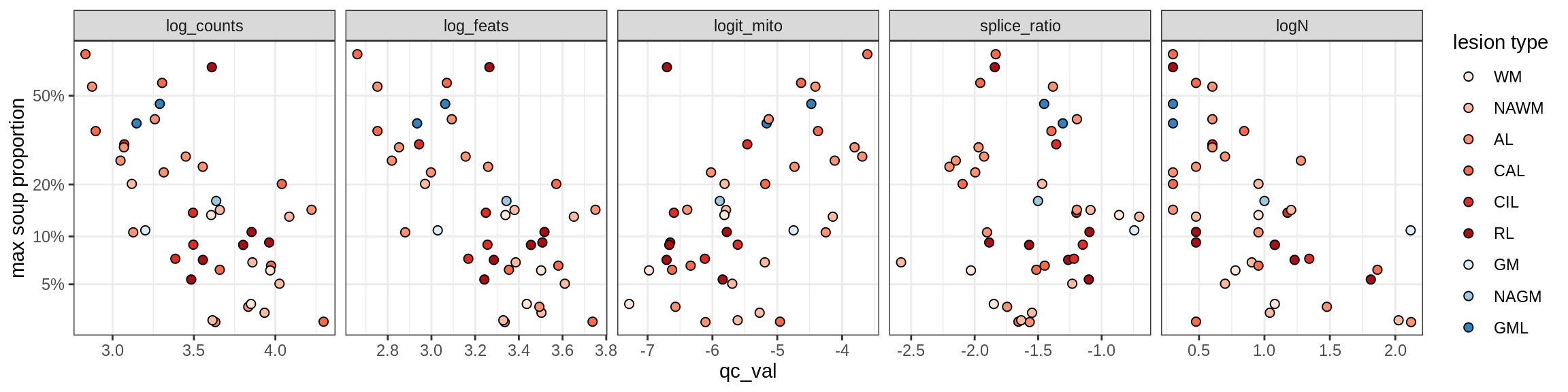
Oligo_A1
Oligo_A2
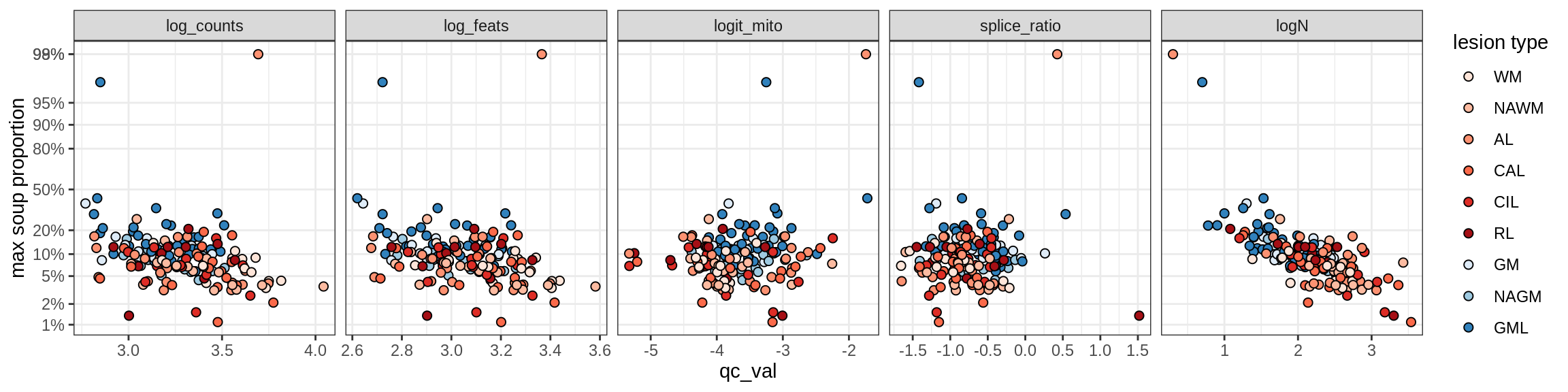
Oligo_B1
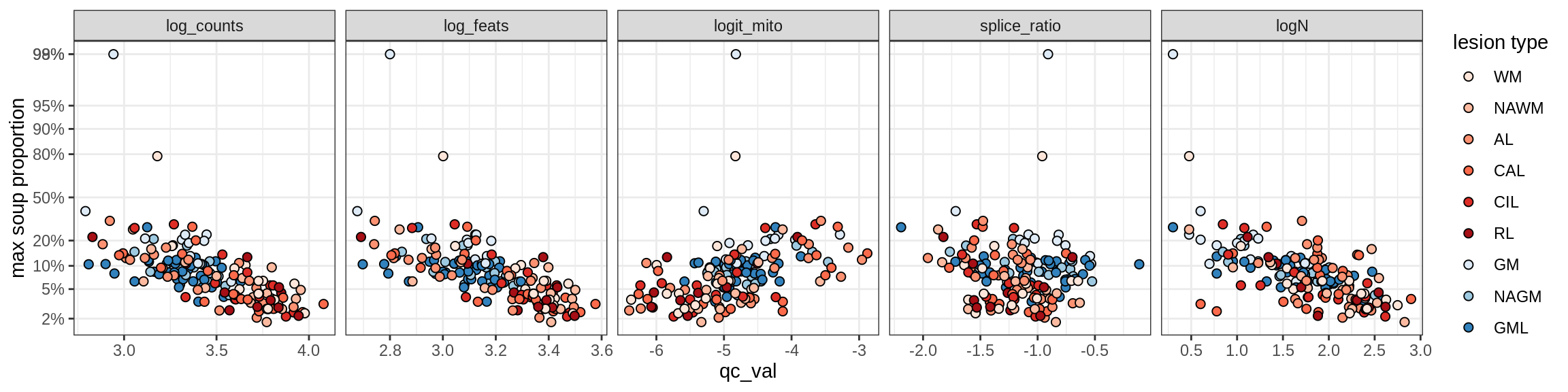
Oligo_B2
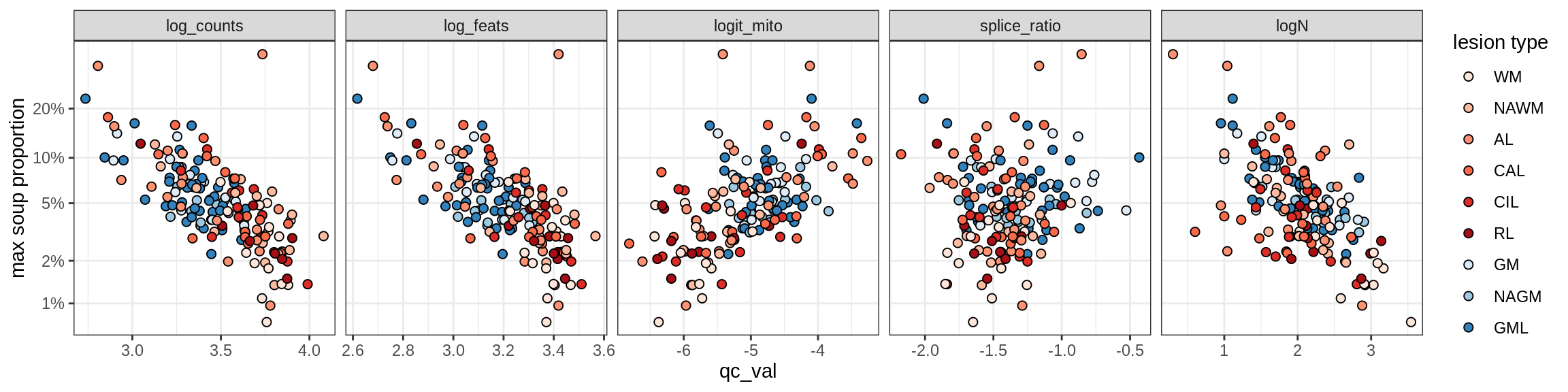
Oligo_B3
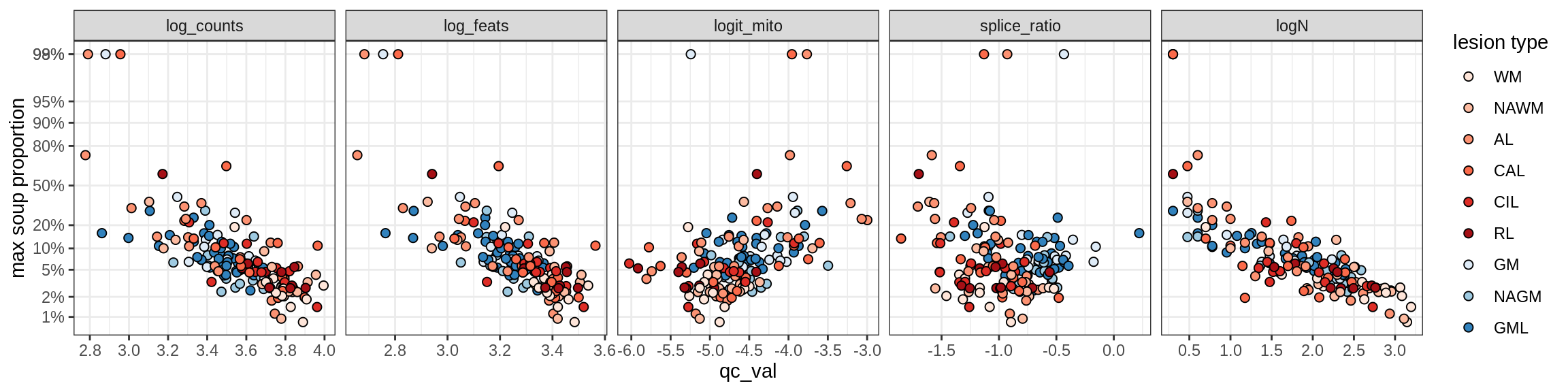
Oligo_B4
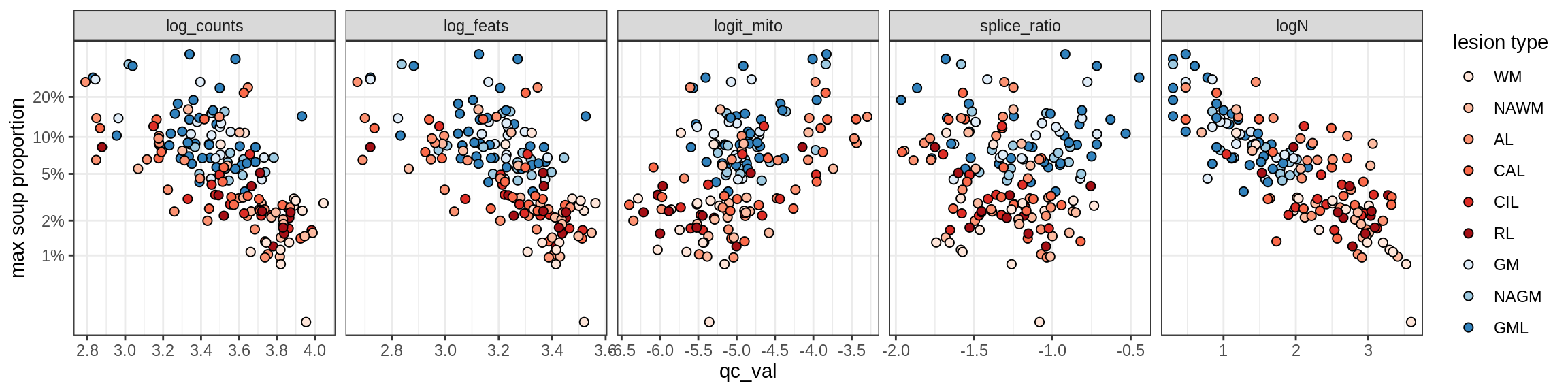
Oligo_C1
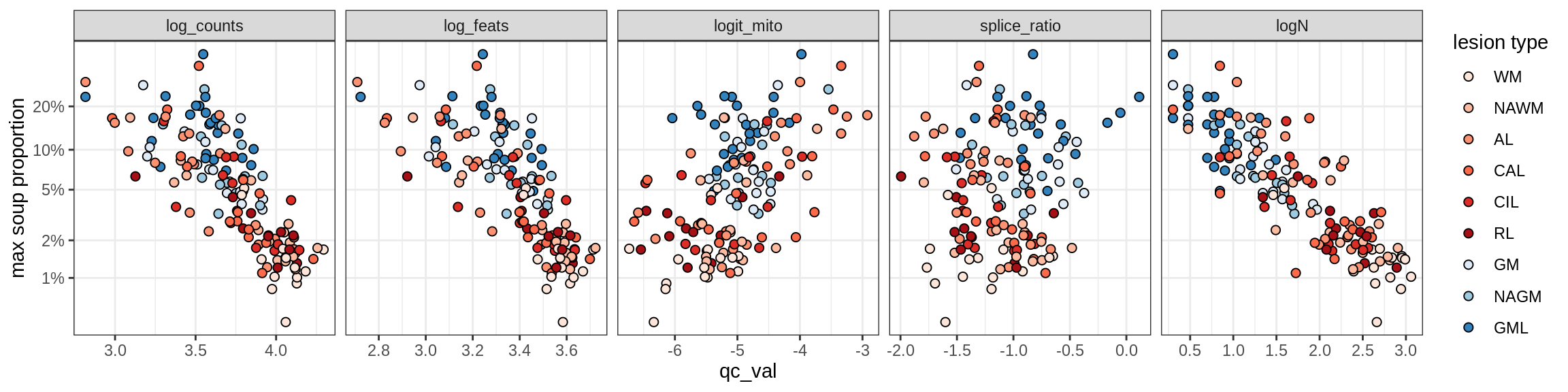
Oligo_C2
Oligo_D
Astrocytes
Astro_A
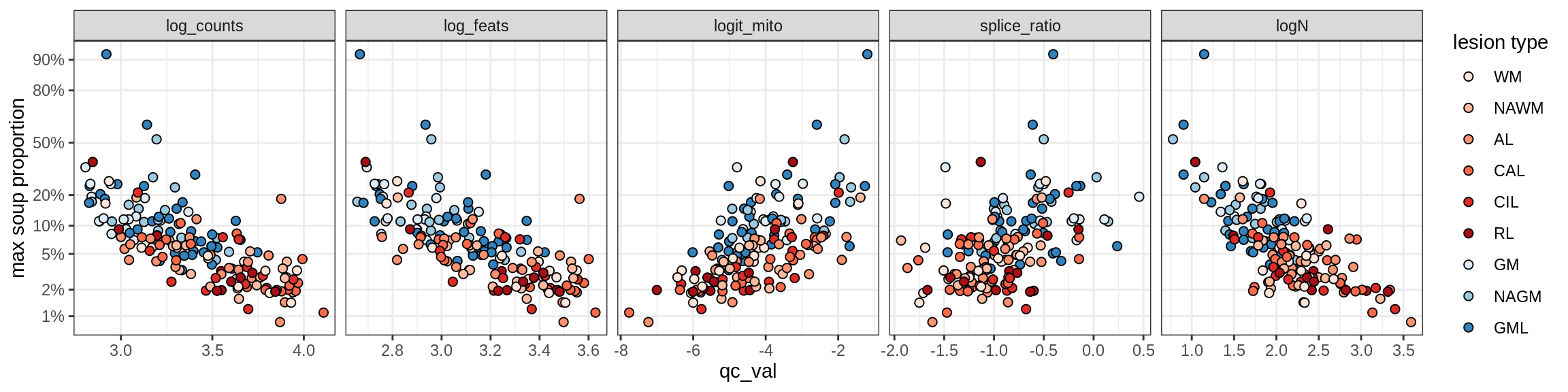
Astro_B
Astro_C
Astro_D
Astro_E
Microglia
Microglia_A
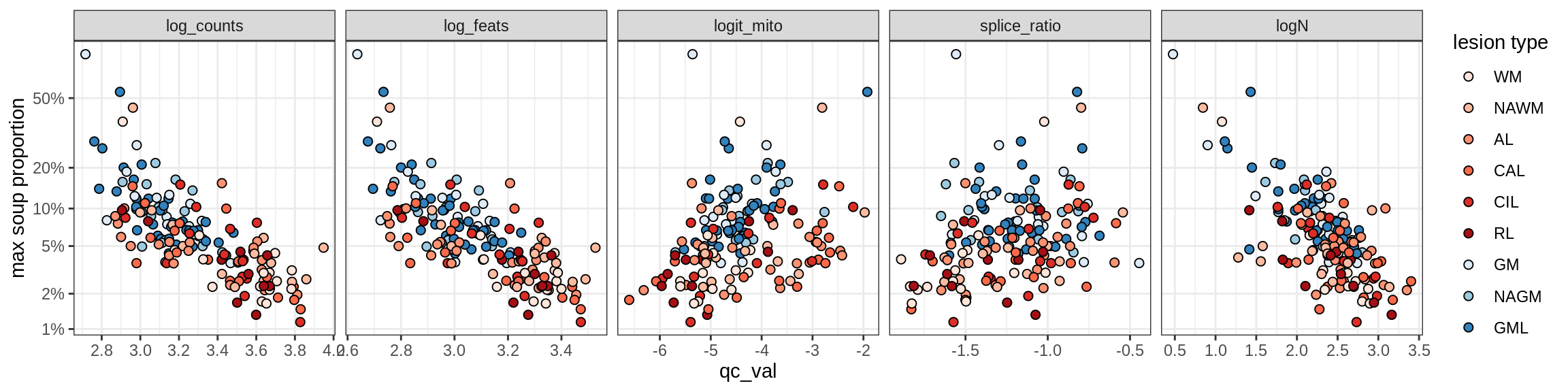
Microglia_B
Excitatory neurons
Ex_CUX2_A
Ex_CUX2_B
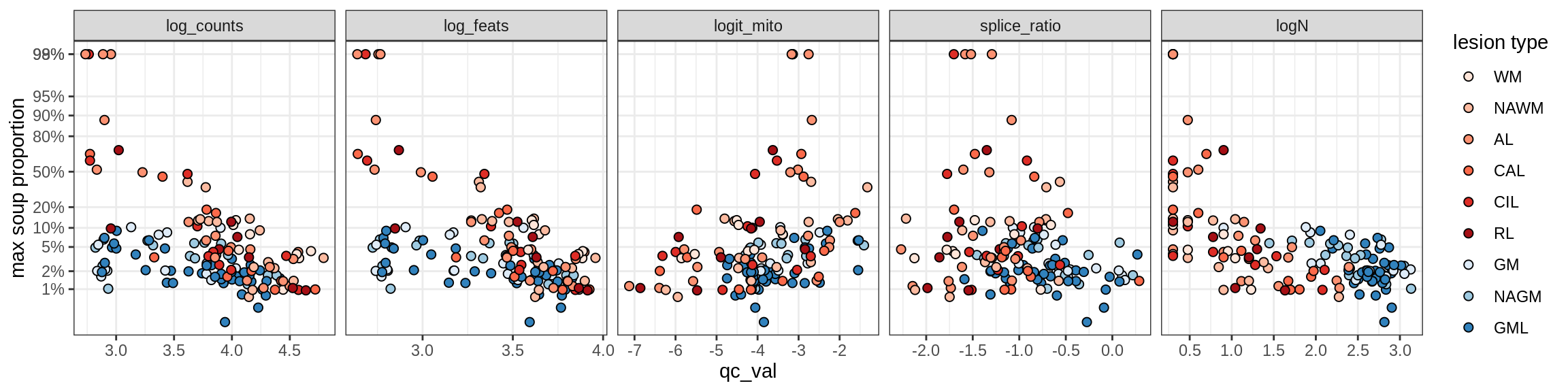
Ex_RORB_A
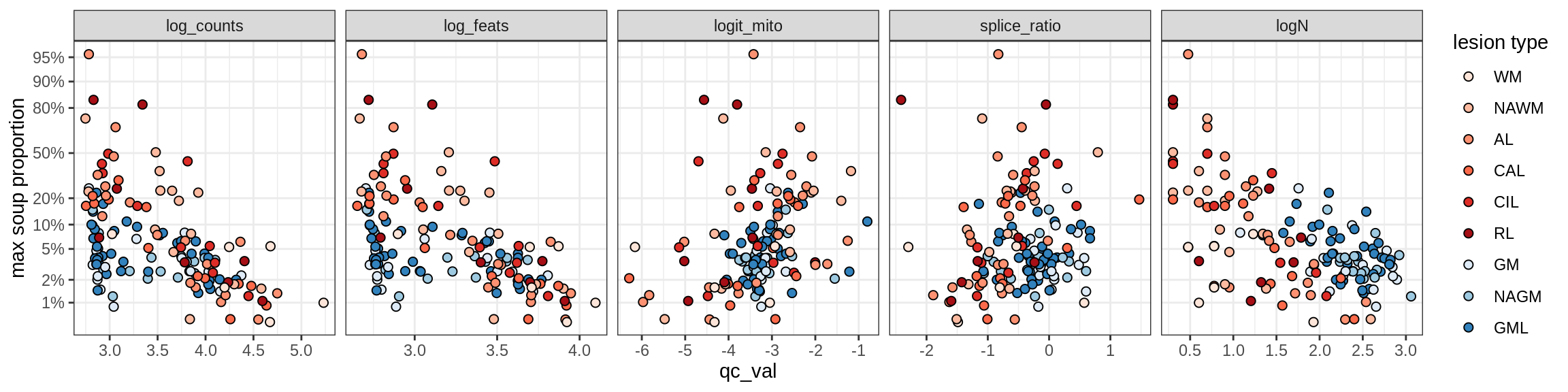
Ex_RORB_B
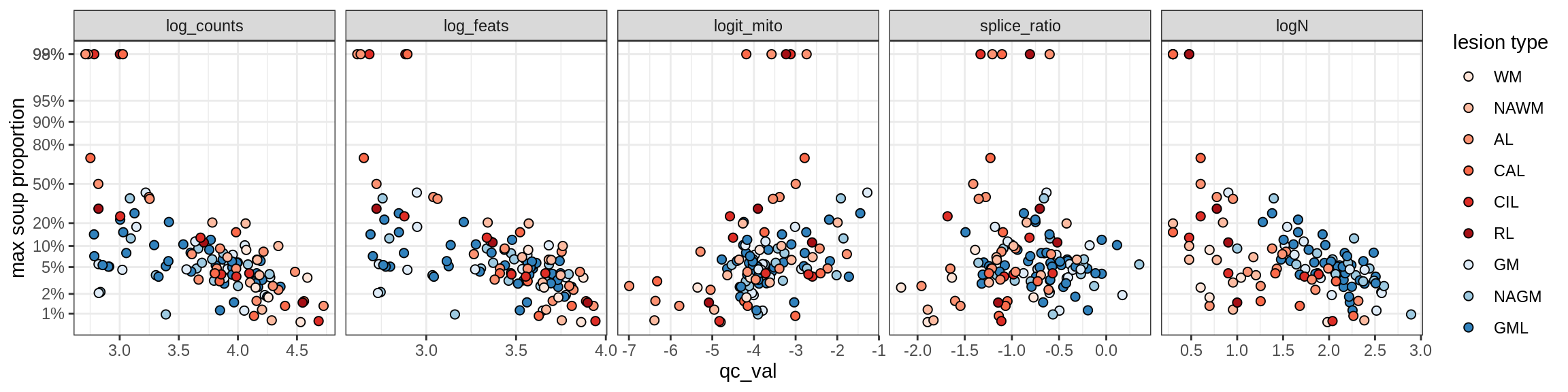
Ex_RORB_C
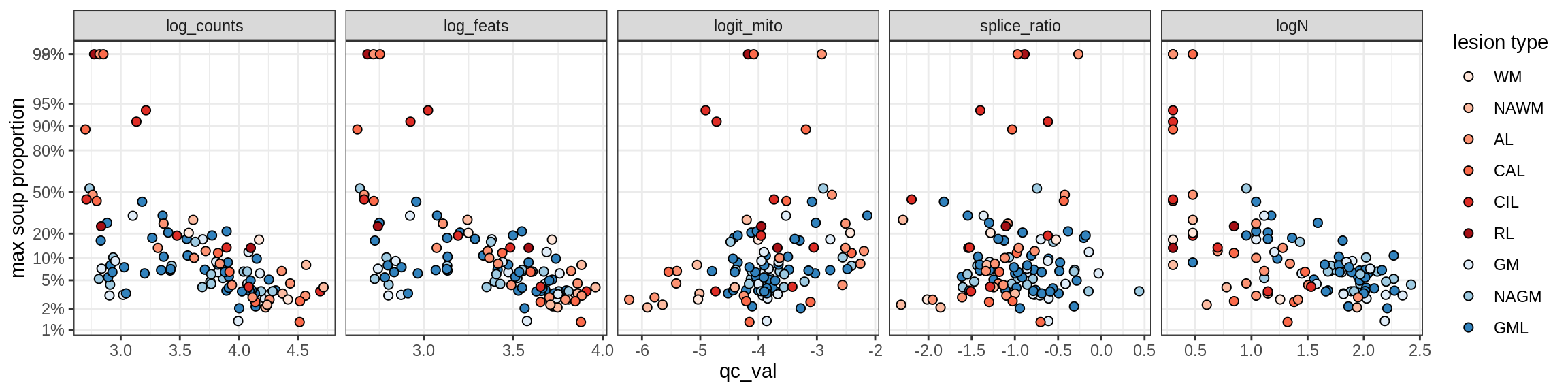
Ex_RORB_CUX2_A
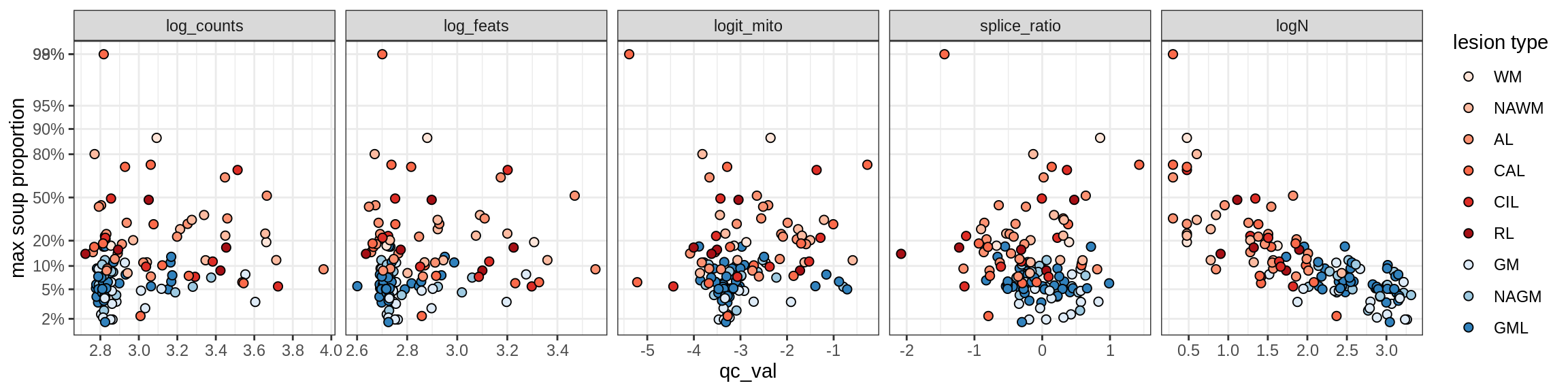
Ex_RORB_CUX2_B
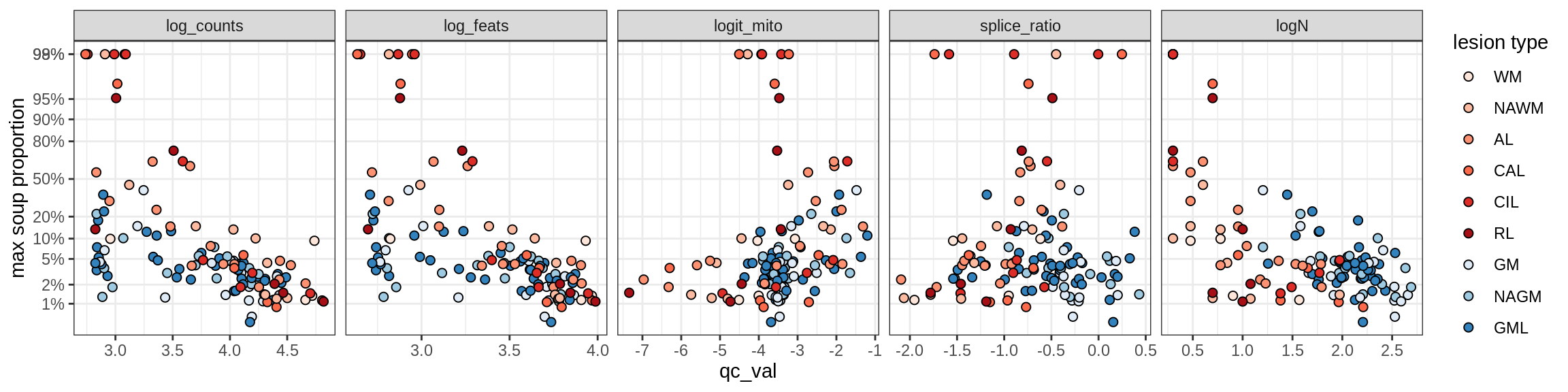
Ex_RORB_CUX2_C
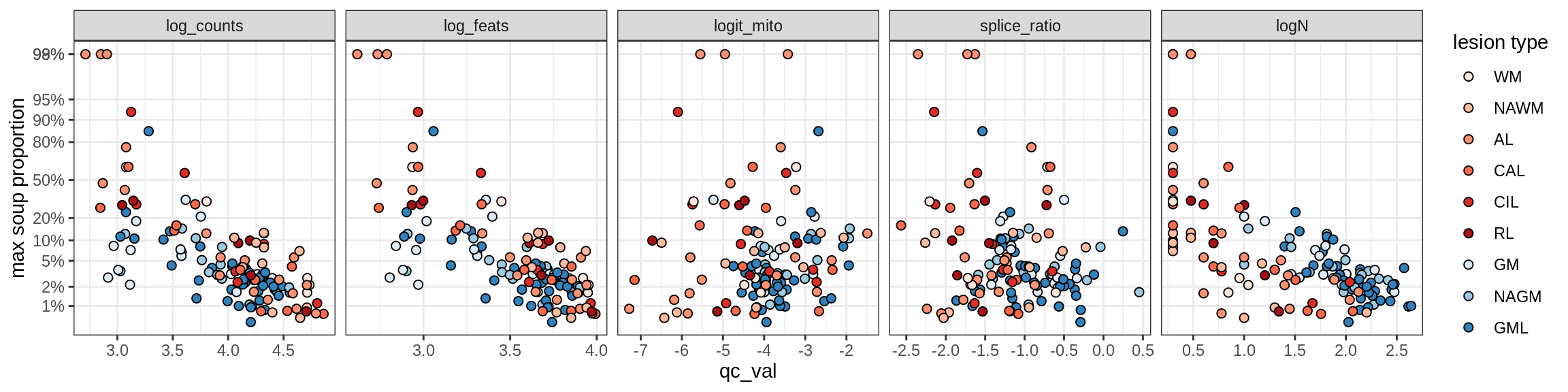
Ex_RORB_CUX2_D
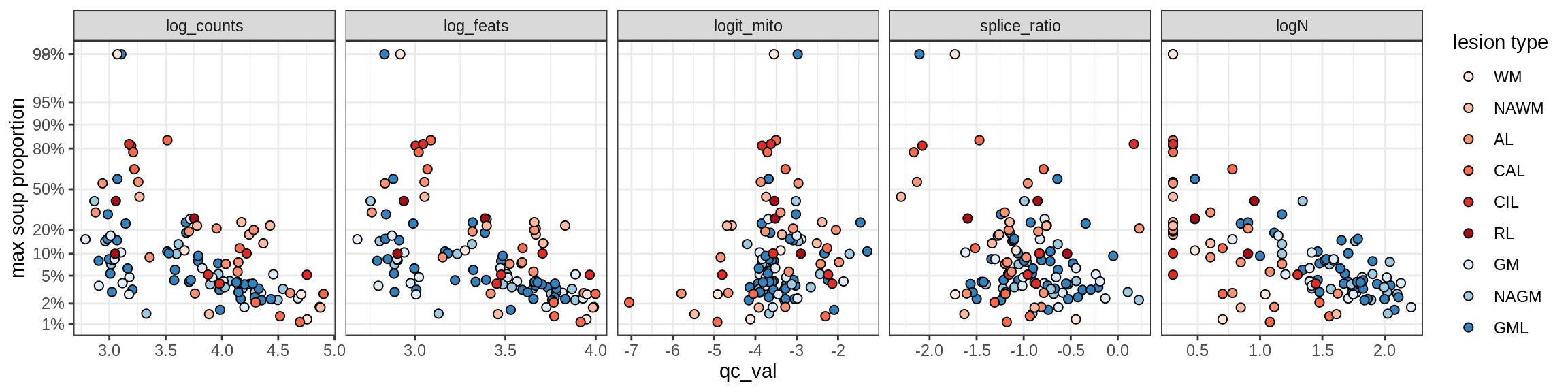
Ex_RORB_CUX2_E
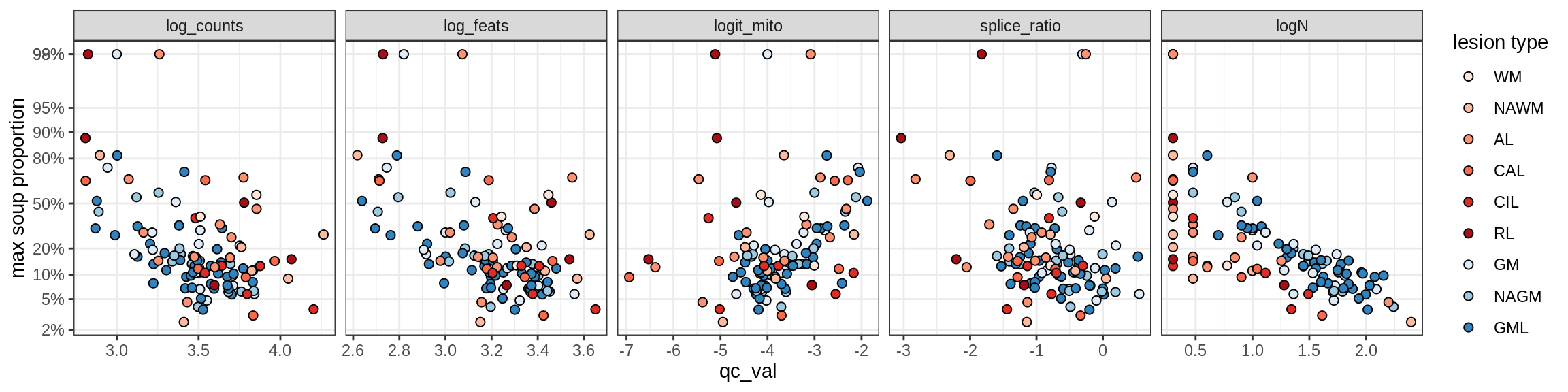
Ex_RORB_CUX2_F
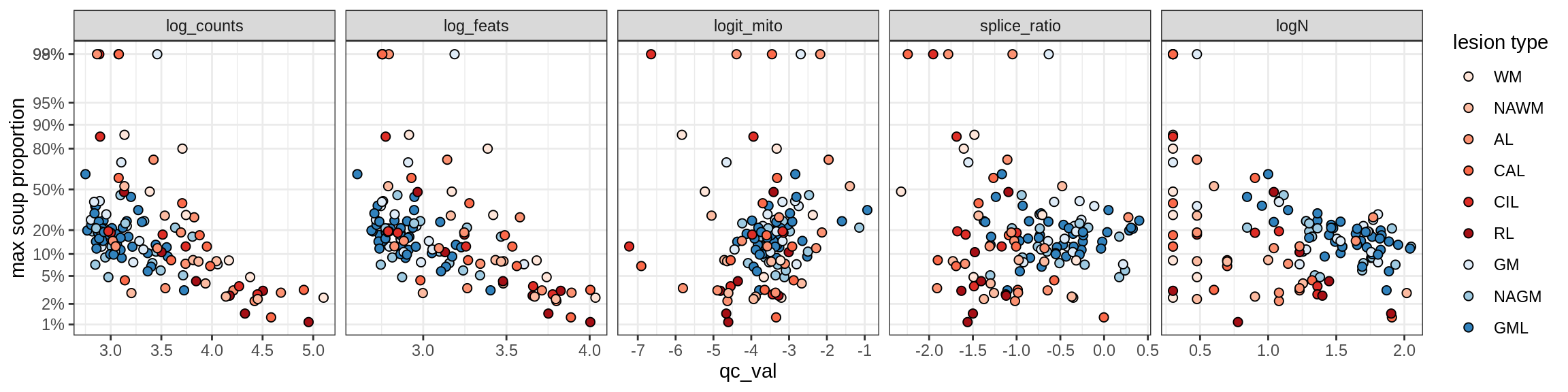
Ex_RORB_THEMIS_A
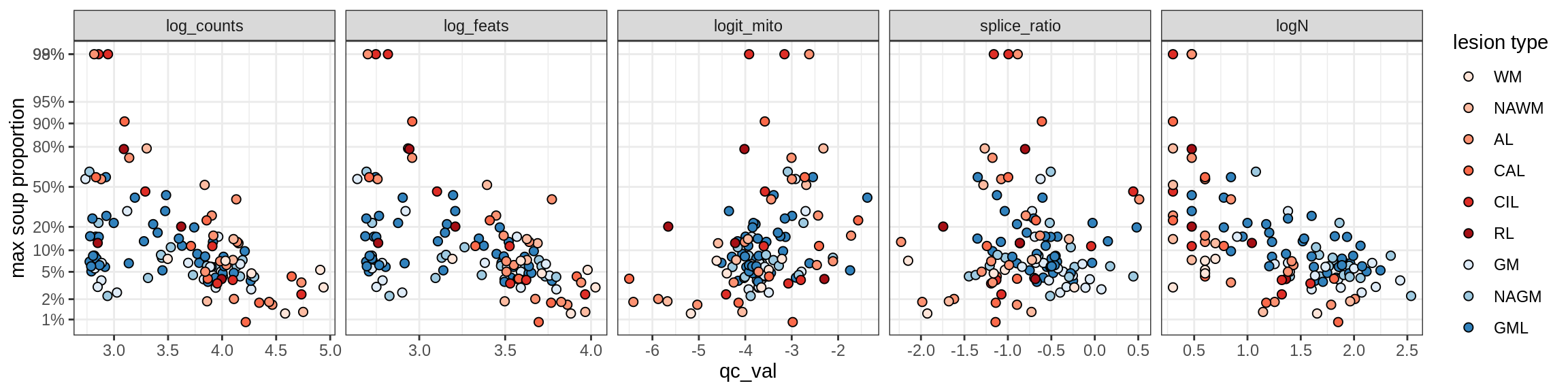
Ex_RORB_THEMIS_B
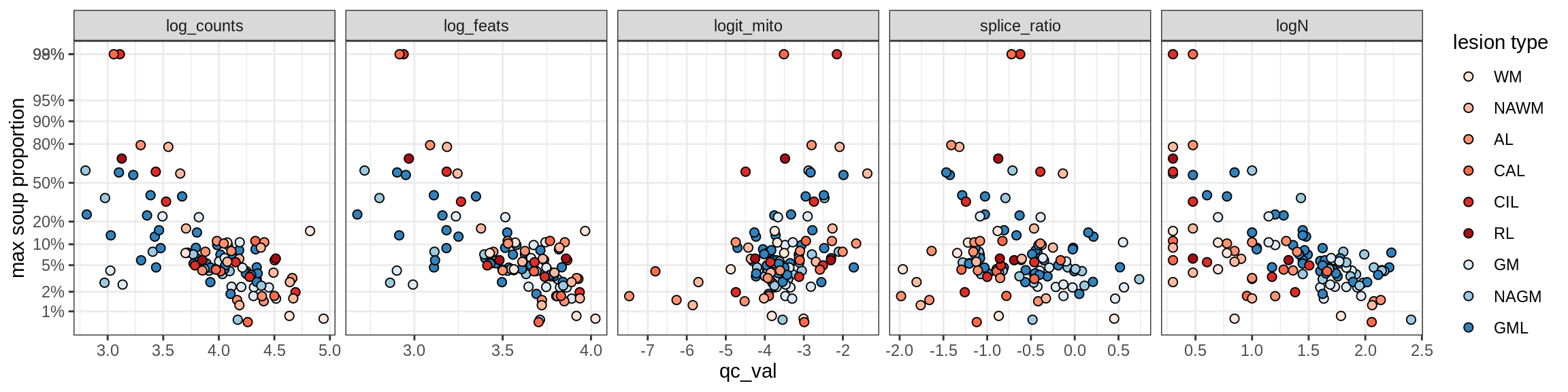
Ex_THEMIS_A
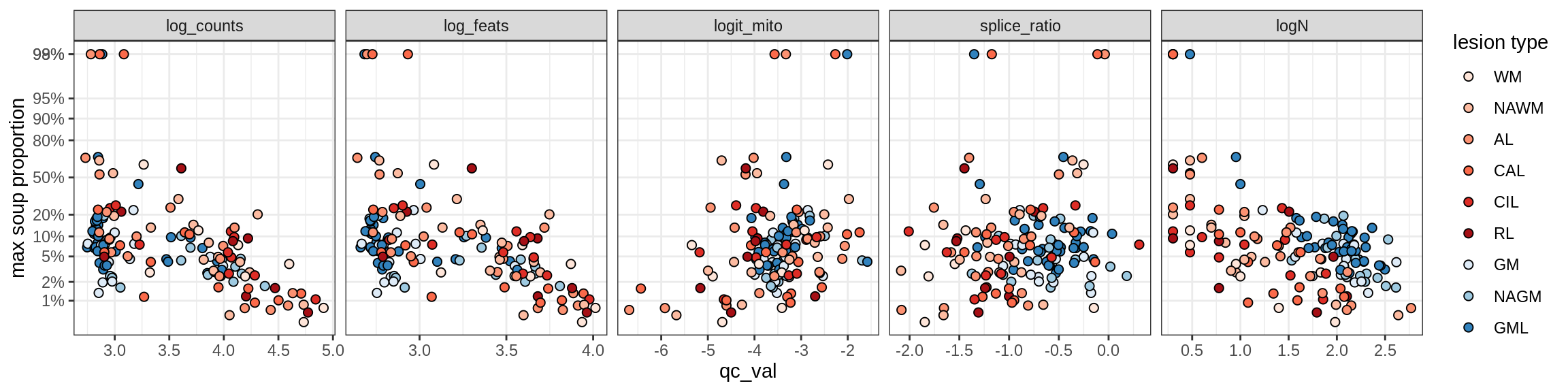
Ex_THEMIS_B
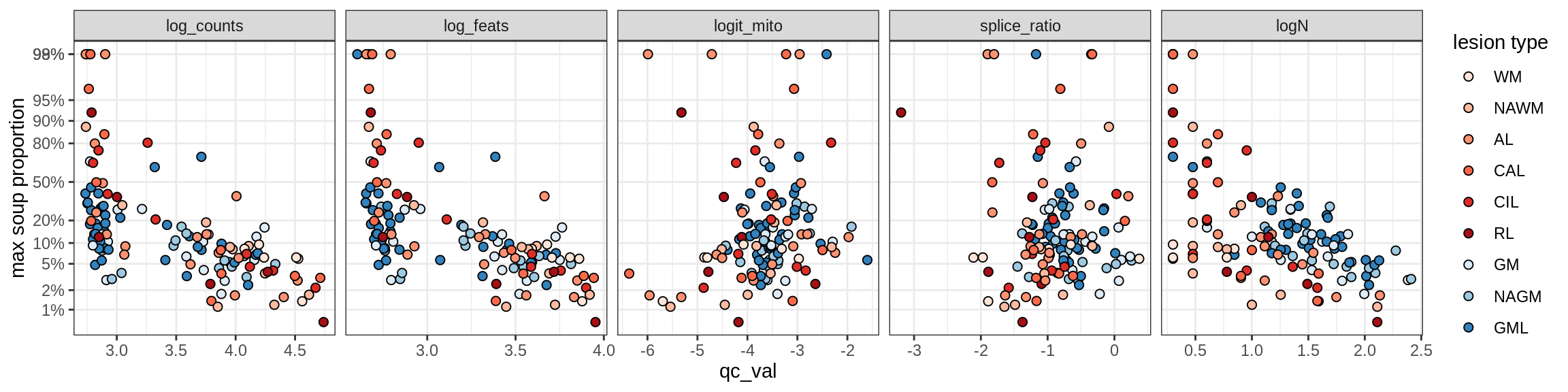
Ex_TLE4_A
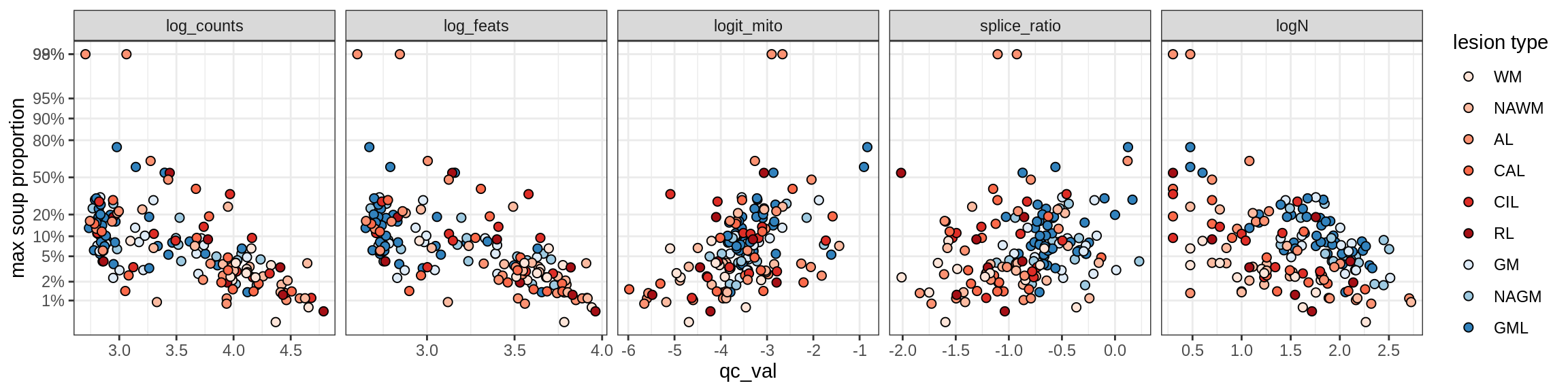
Ex_TLE4_B
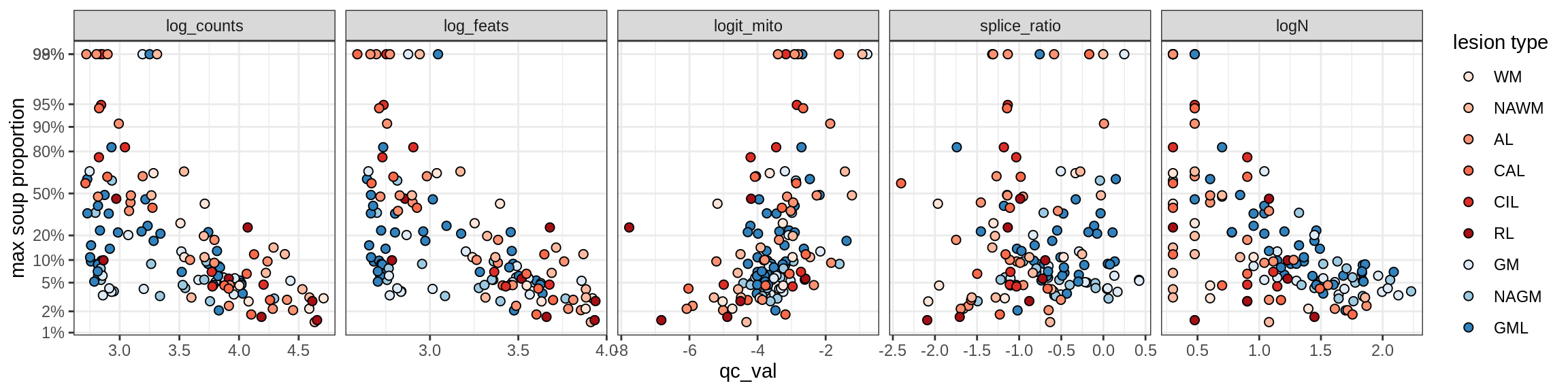
Neuro_oligo
Inhibitory neurons
Inh_LAMP5
Inh_Pvalb_A
Inh_Pvalb_B
Inh_RELN
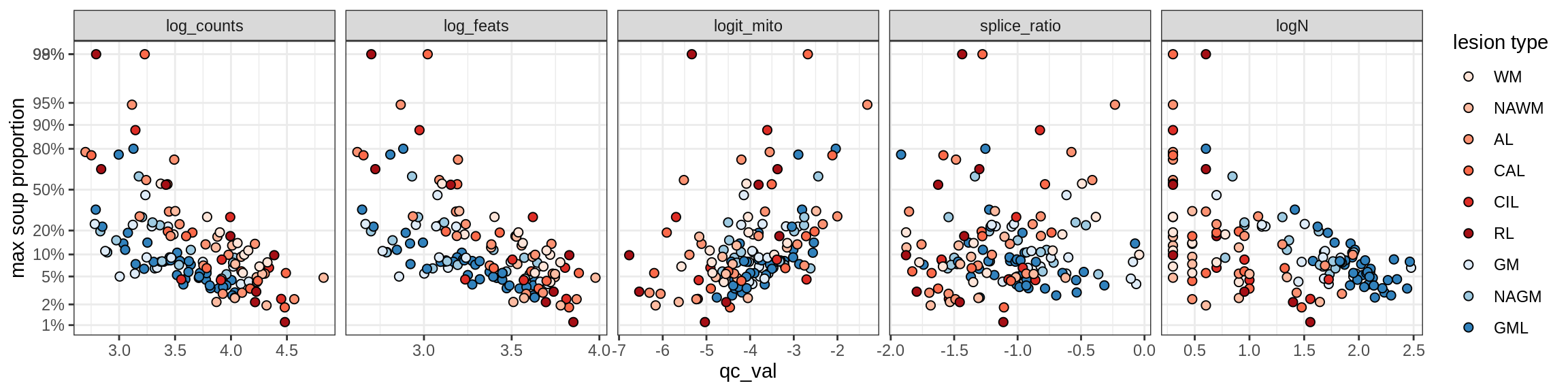
Inh_SST_A
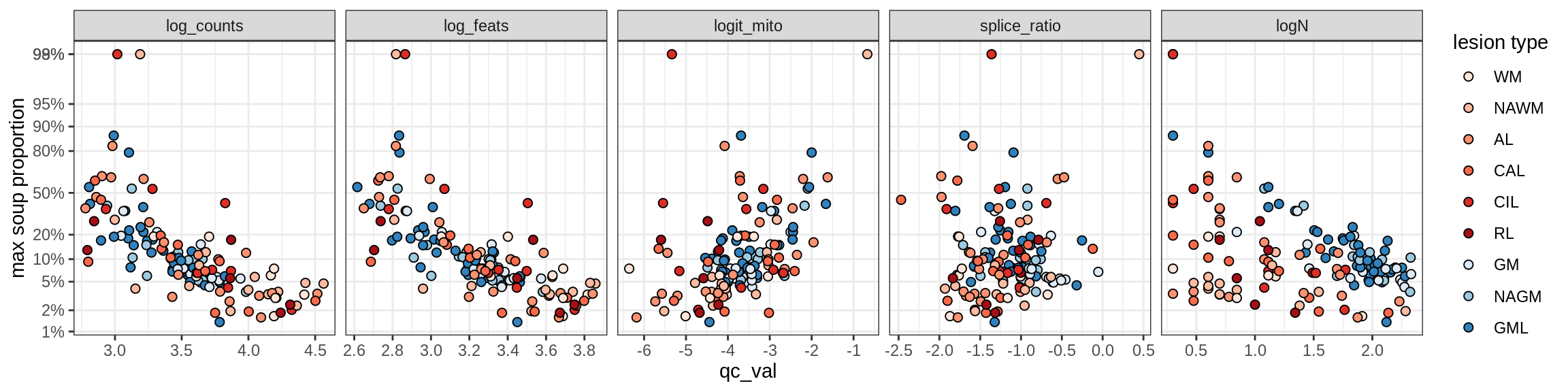
Inh_SST_B
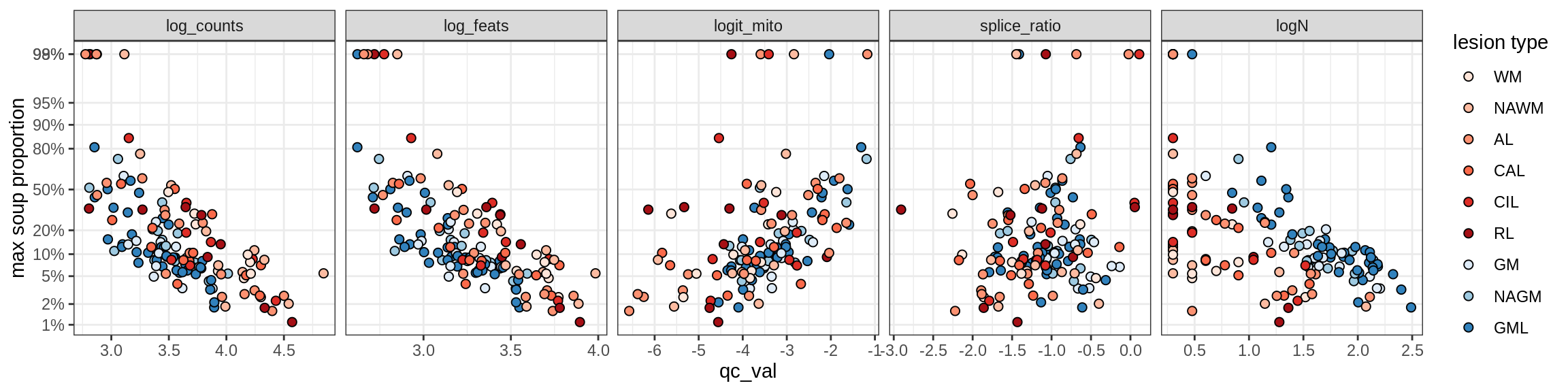
Inh_VIP
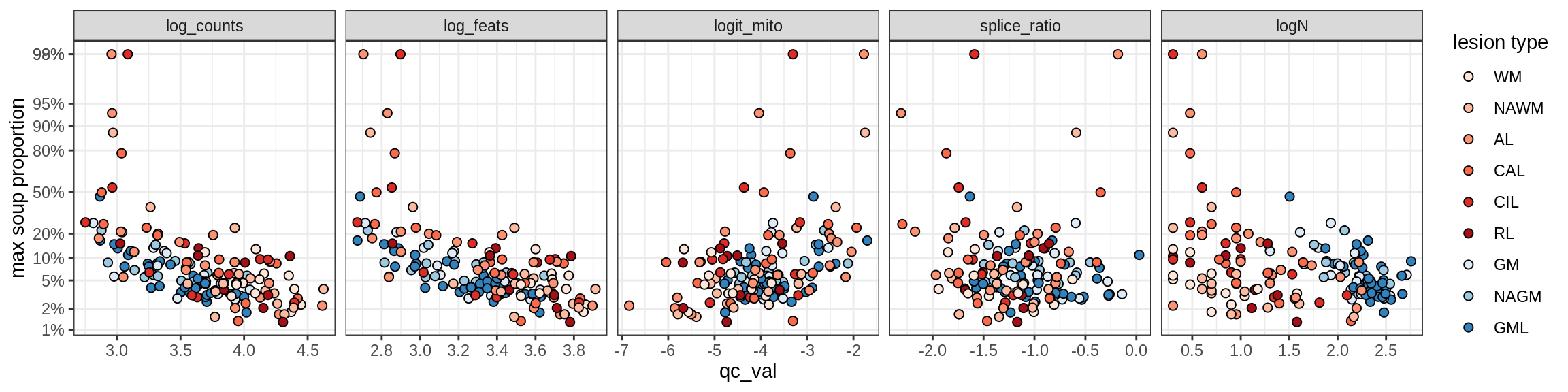
Endothelial cells
Endo_A
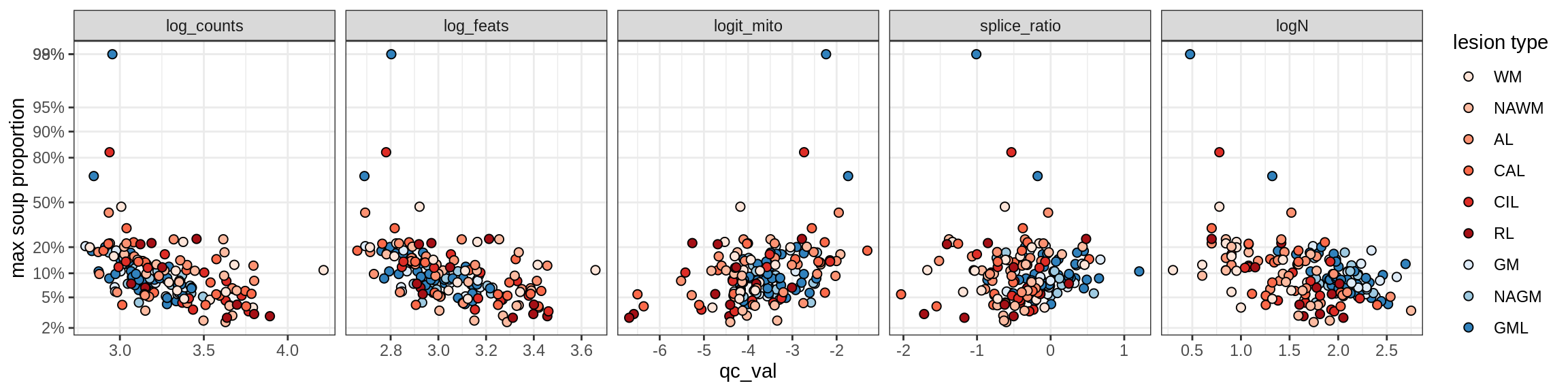
Endo_B
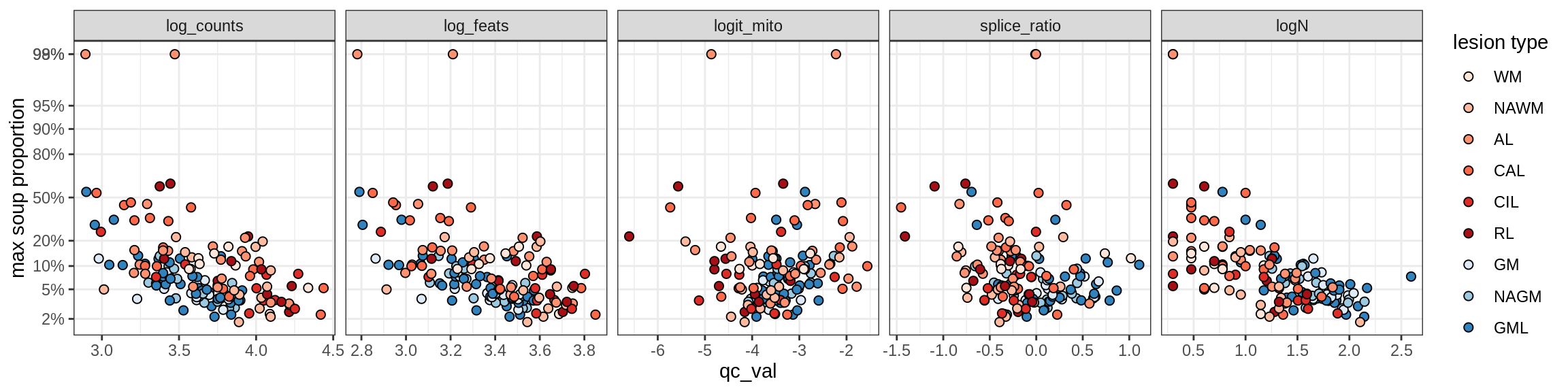
Pericytes
Pericytes
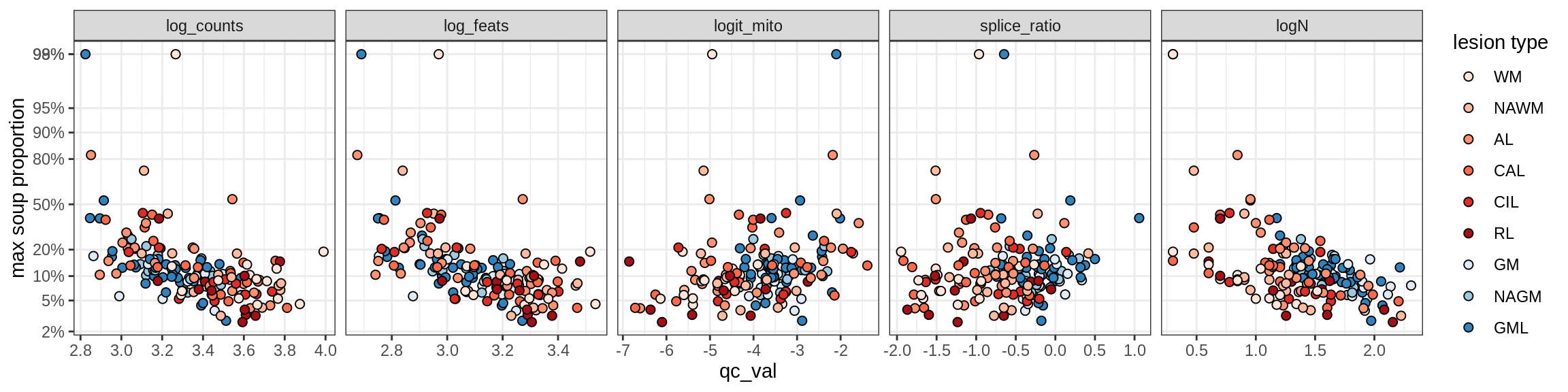
Immune
B_cells
T_cells
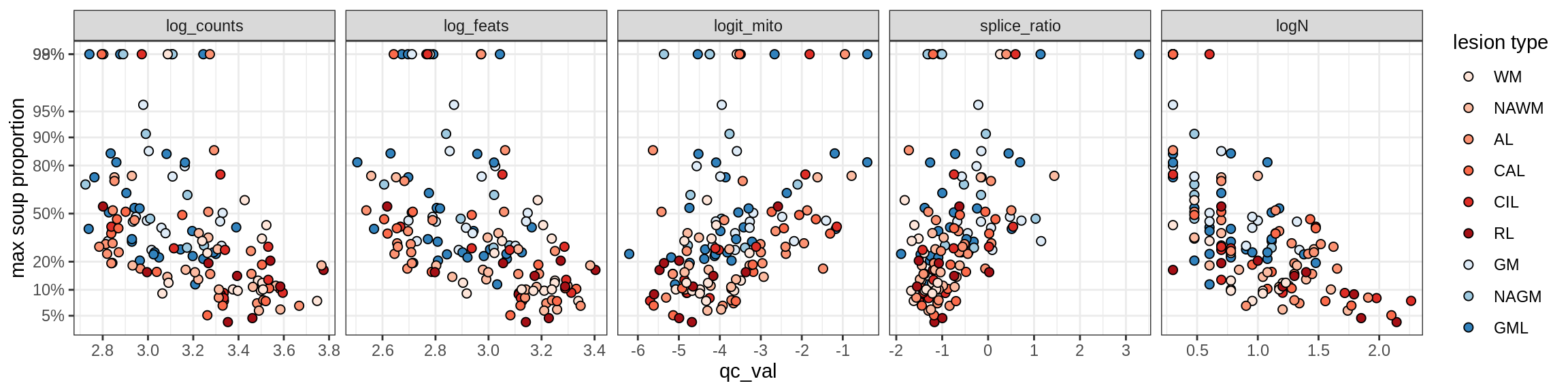
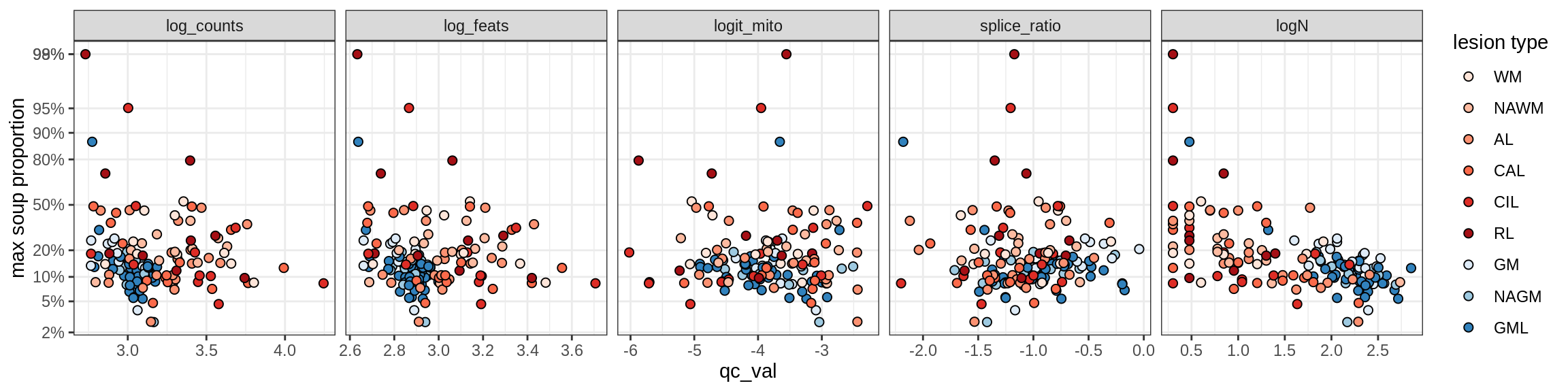
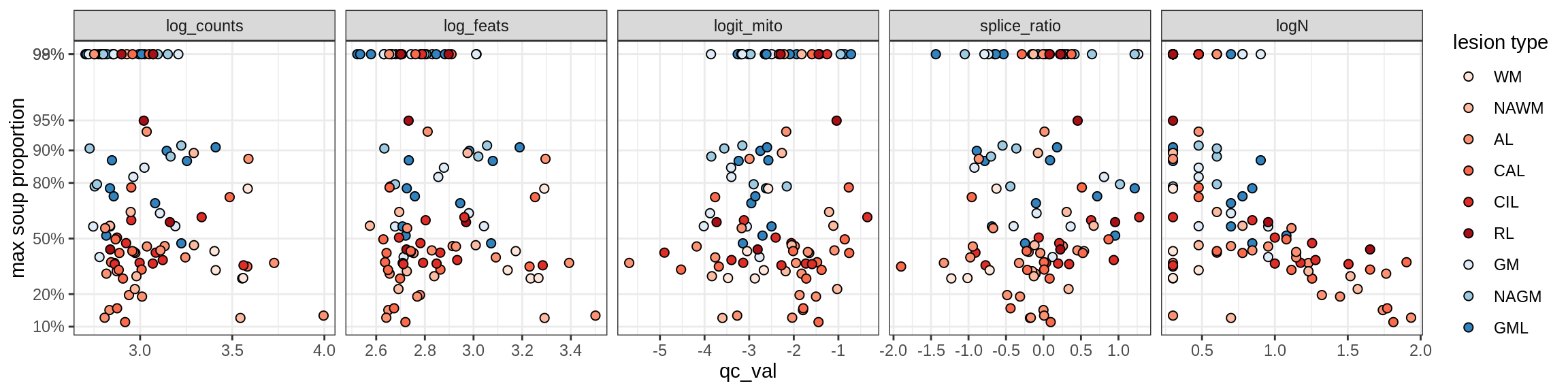
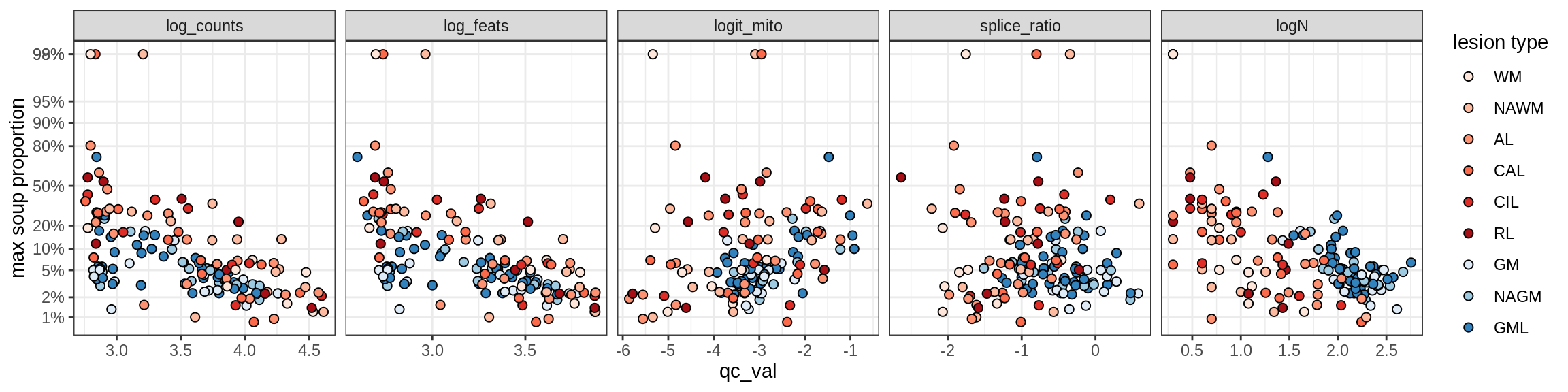
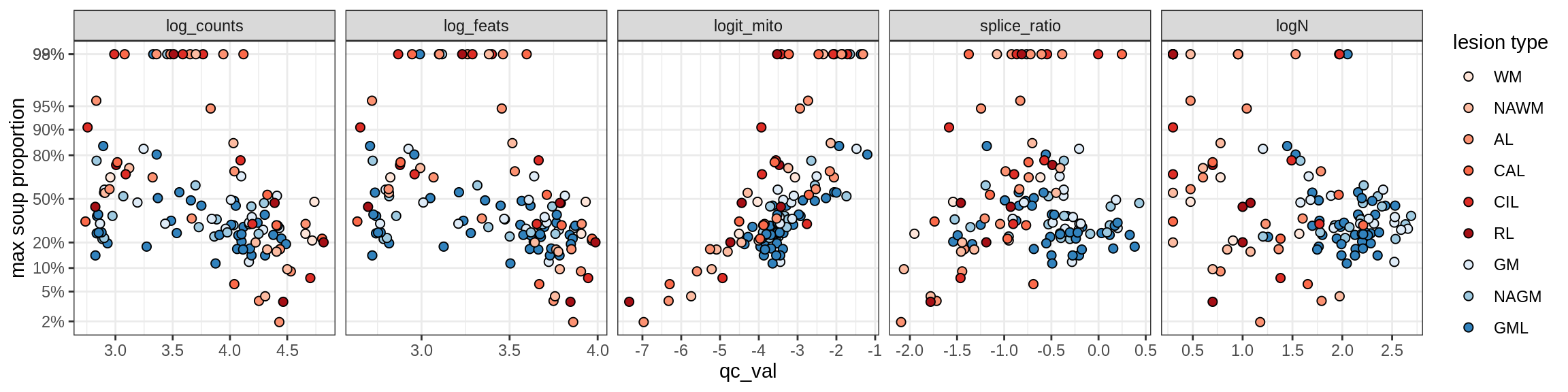
Correlations of mito contamination with QC metrics
for (b in broad_ord) {
cat('### ', b, '{.tabset}\n')
for (t in levels(fct_drop(props_mt[type_broad == b]$type_fine))) {
cat('#### ', t, '\n')
suppressWarnings(print(plot_soup_props_vs_qc(props_mt[type_fine == t], qc_stats)))
cat('\n\n')
}
}OPCs / COPs
OPC
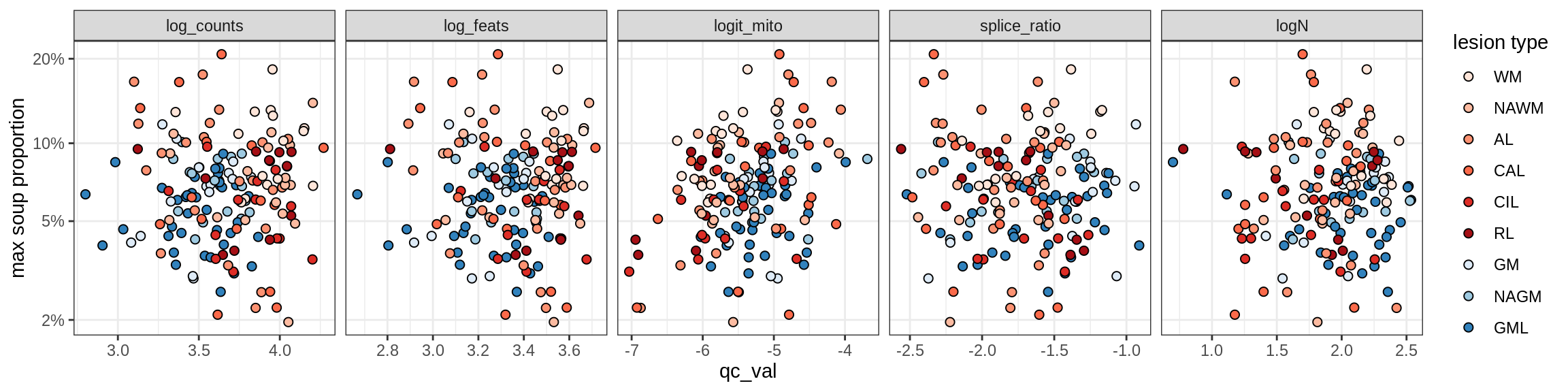
COP_A1
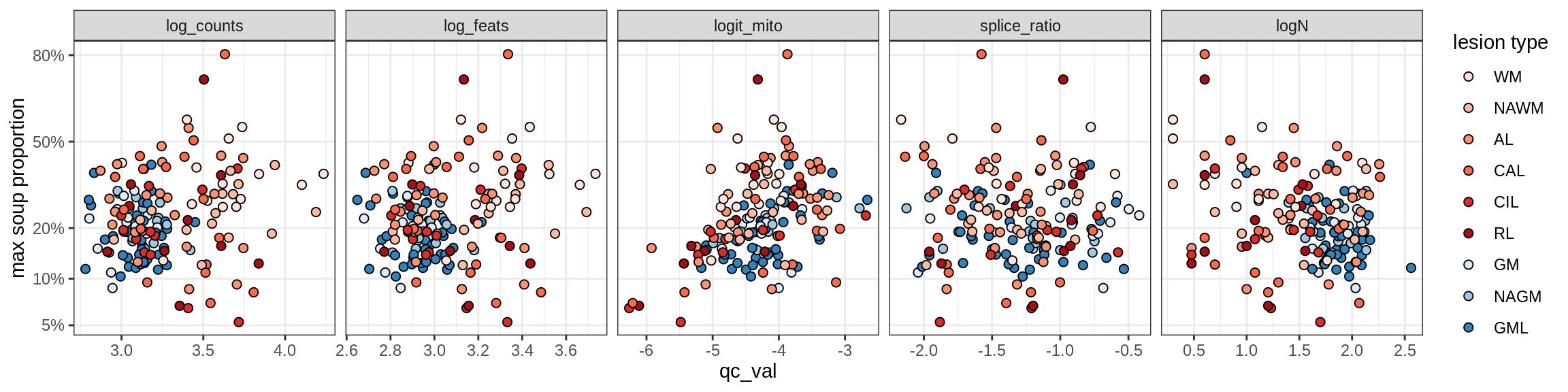
COP_A2
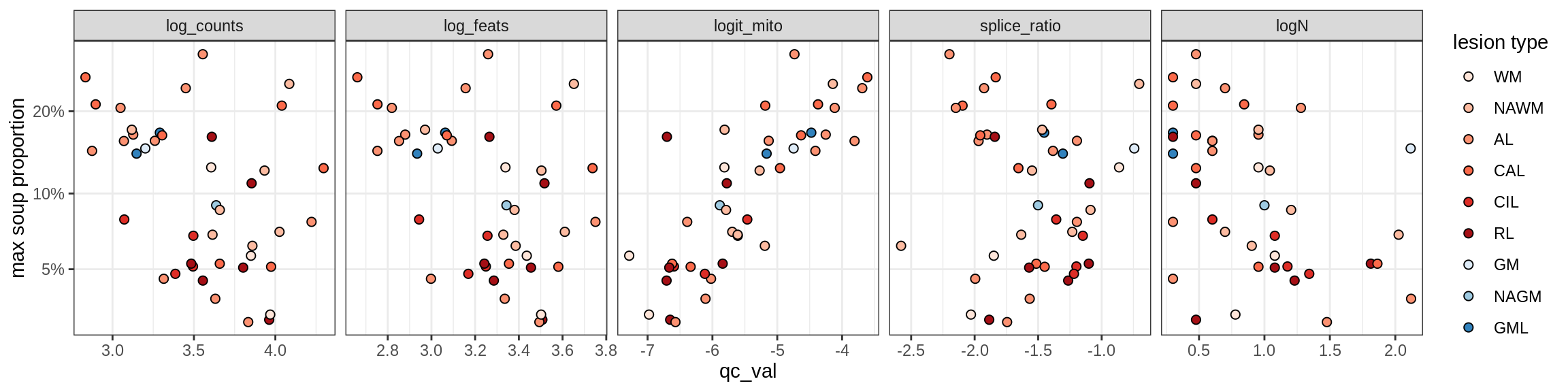
COP_B
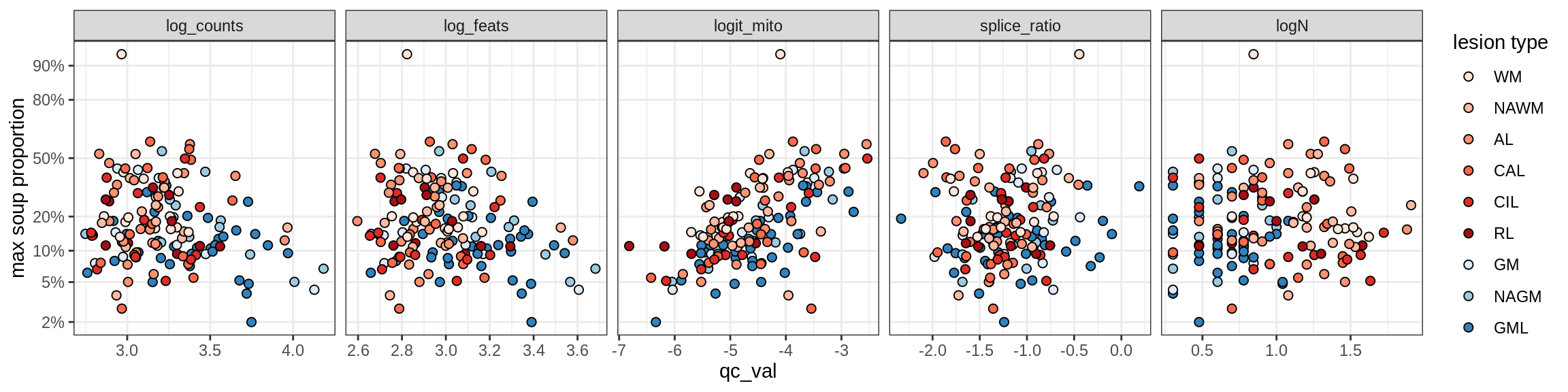
Oligodendrocytes
Oligo_A1
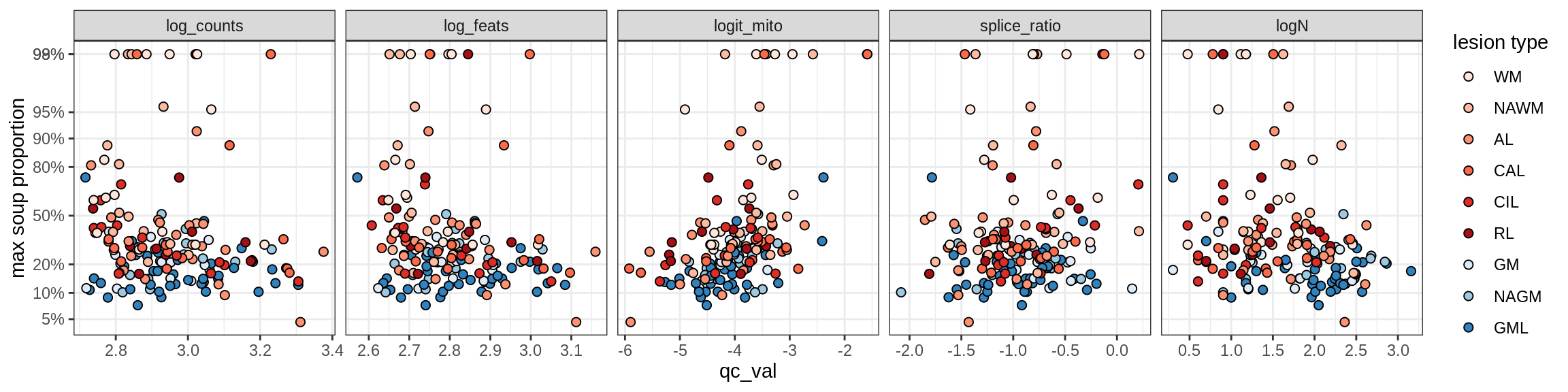
Oligo_A2

Oligo_B1
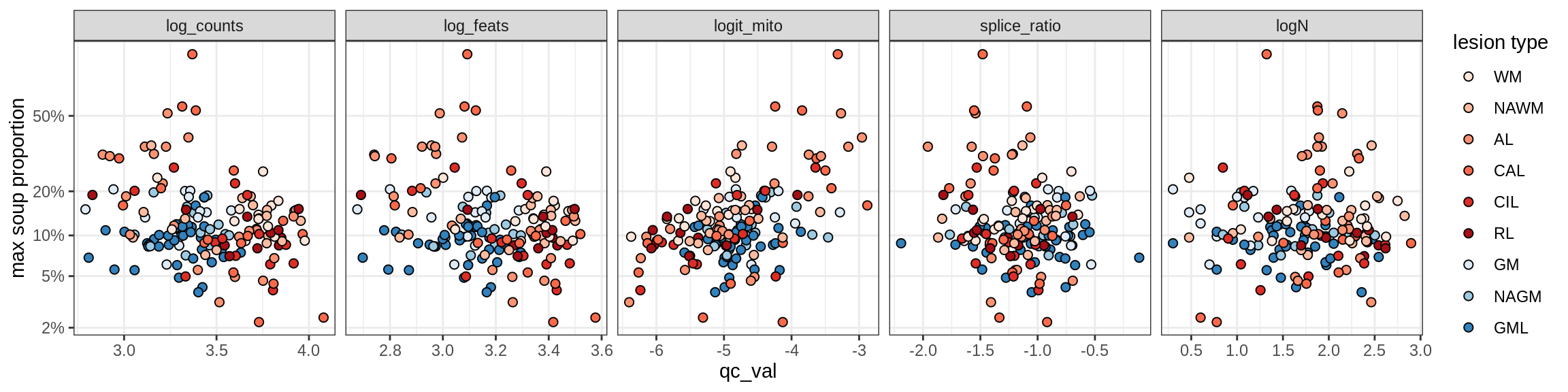
Oligo_B2
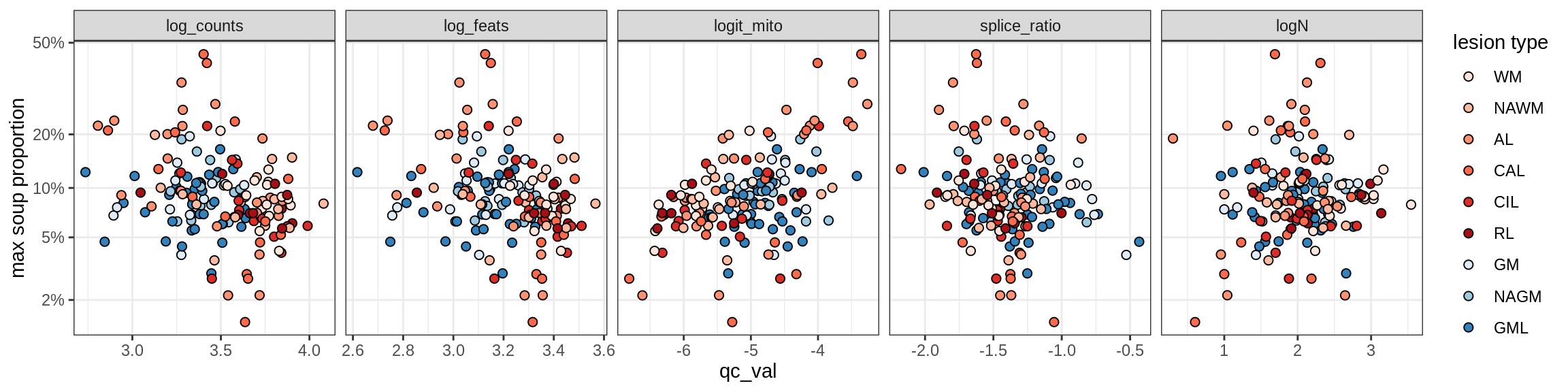
Oligo_B3
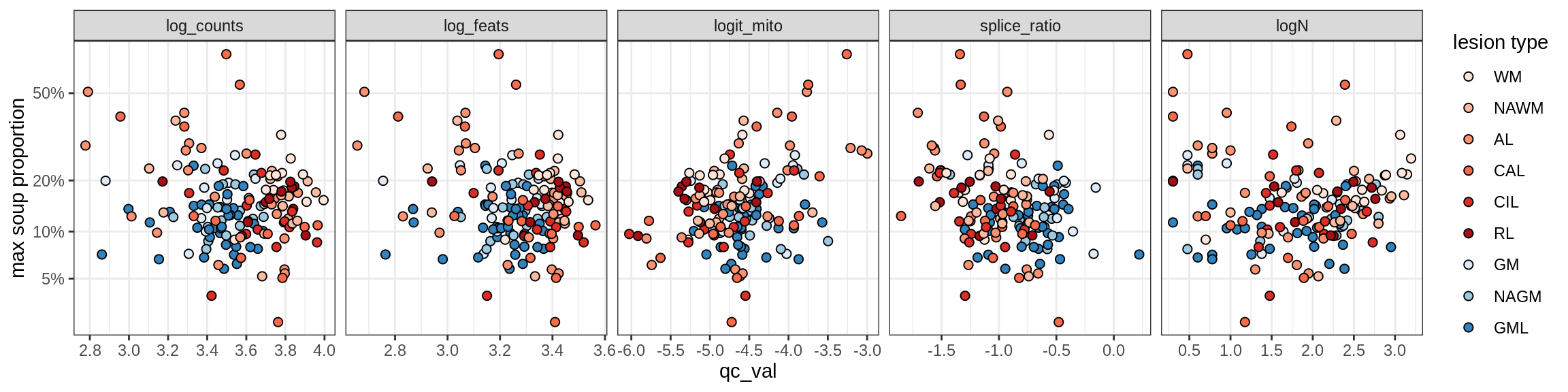
Oligo_B4
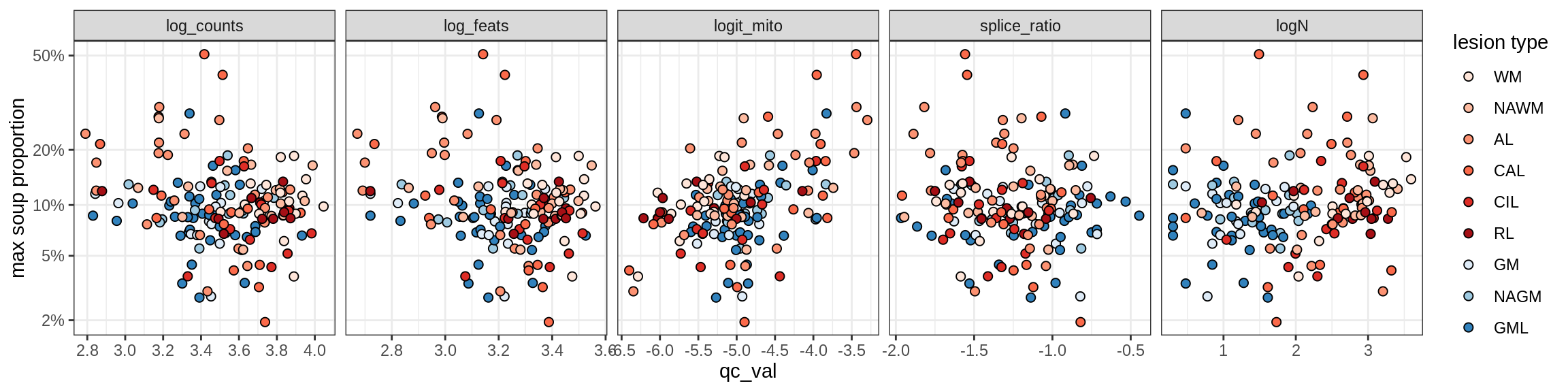
Oligo_C1
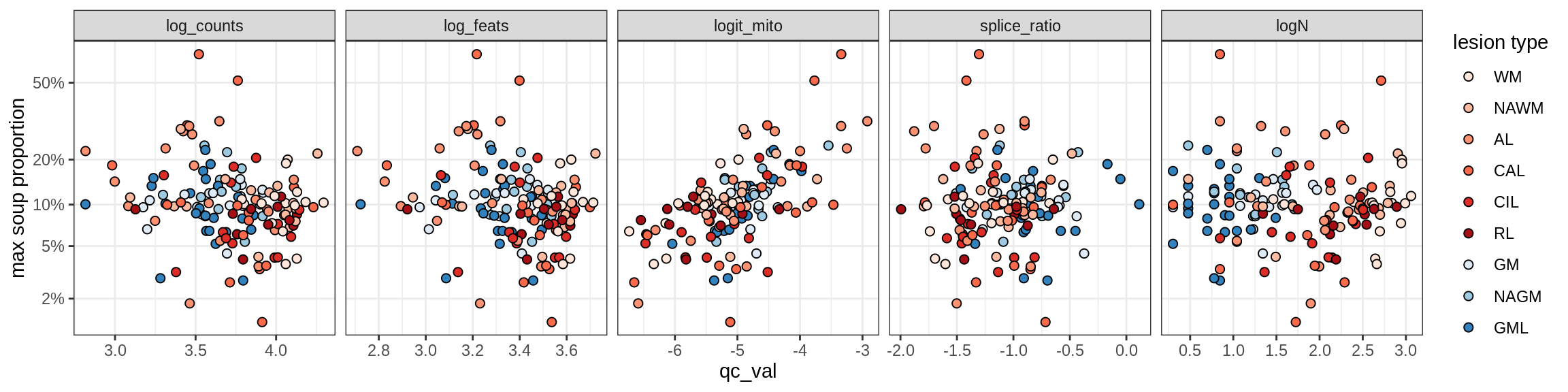
Oligo_C2
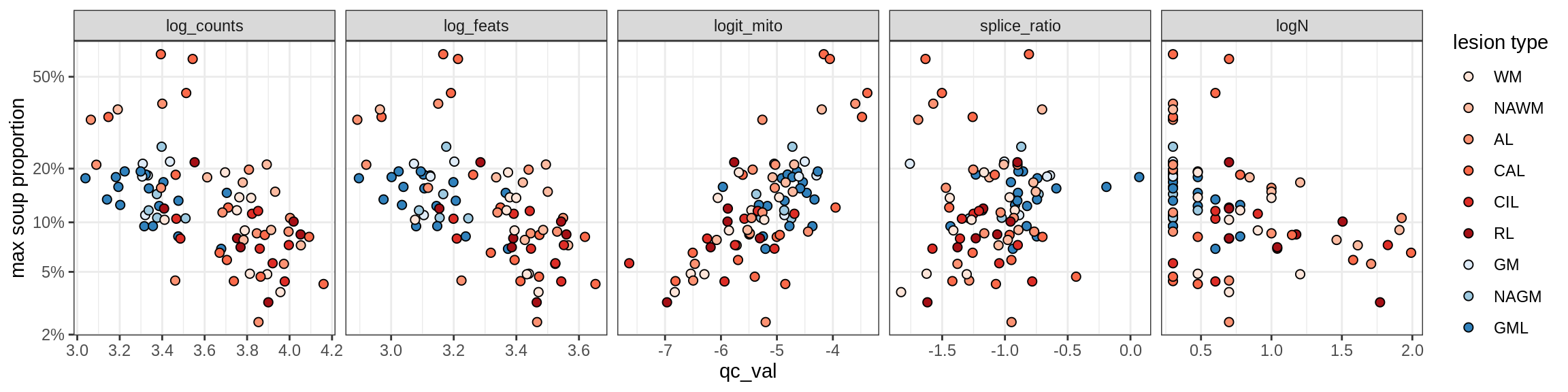
Oligo_D

Astrocytes
Astro_A
Astro_B
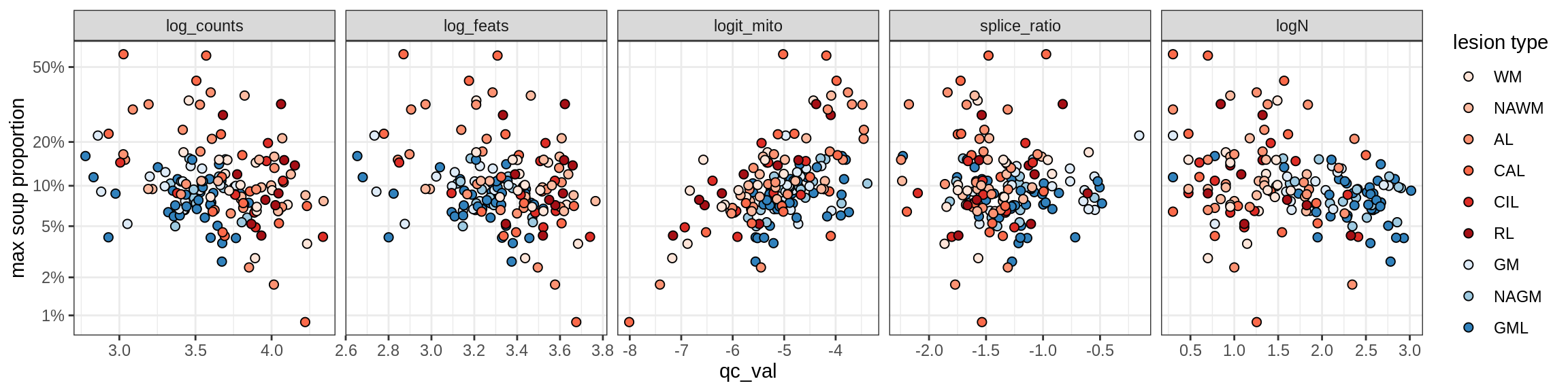
Astro_C
Astro_D
Astro_E
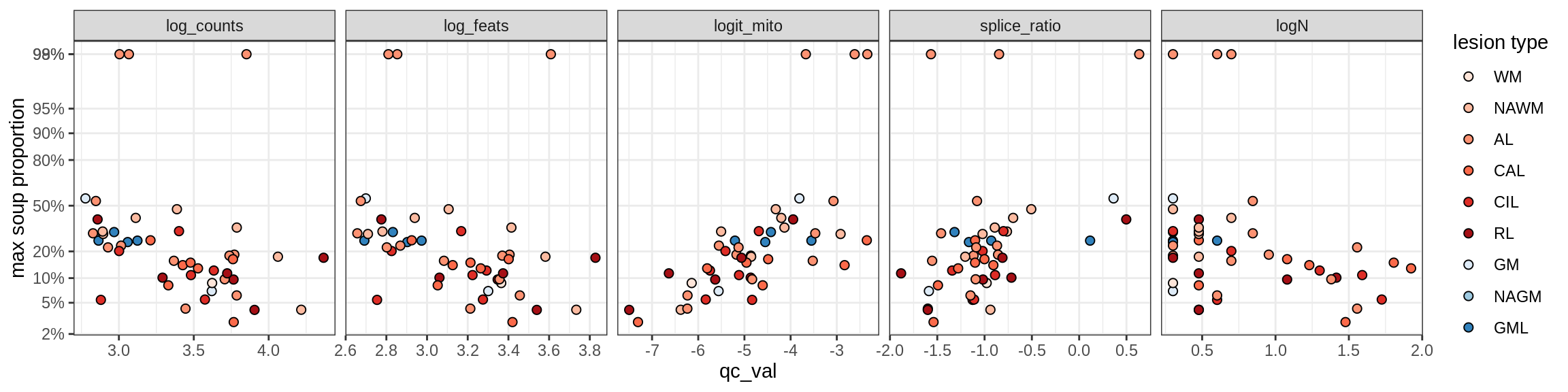
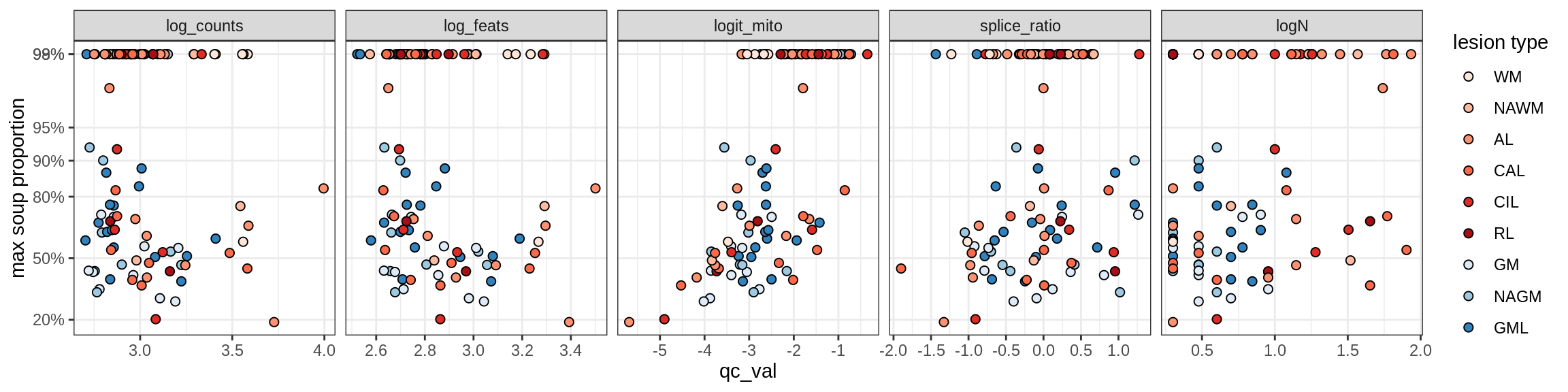
Microglia
Microglia_A
Microglia_B
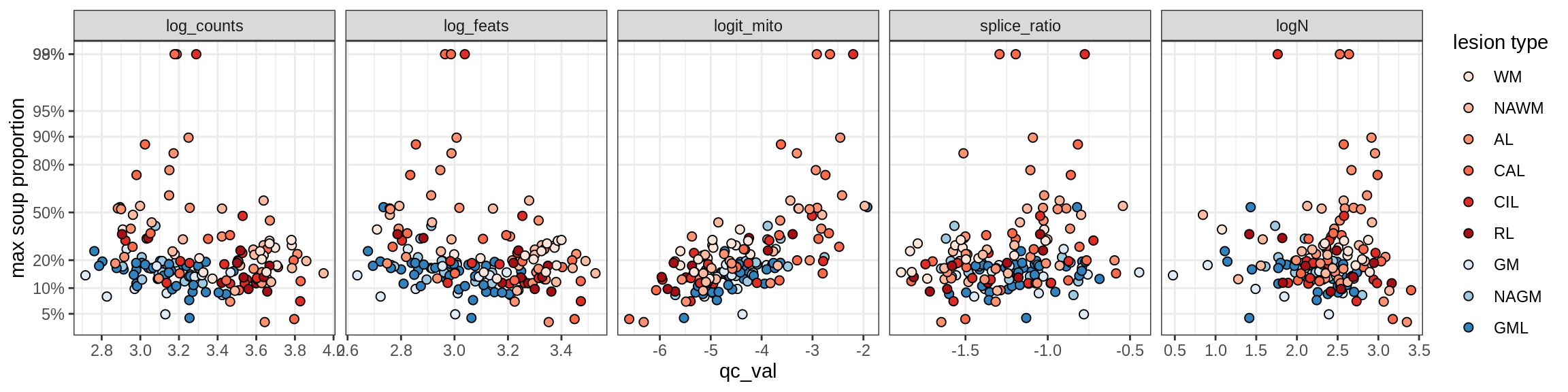
Excitatory neurons
Ex_CUX2_A
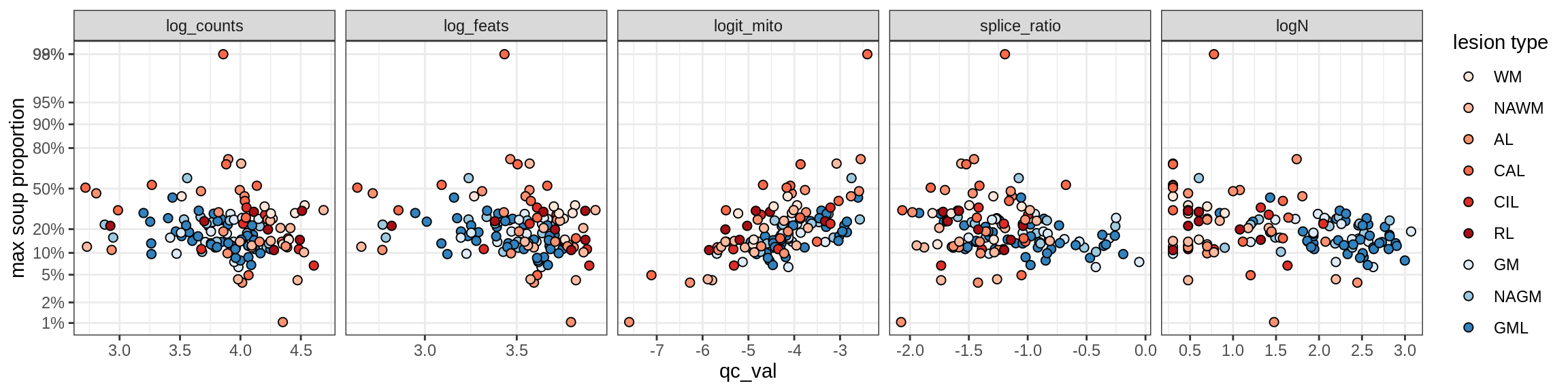
Ex_CUX2_B
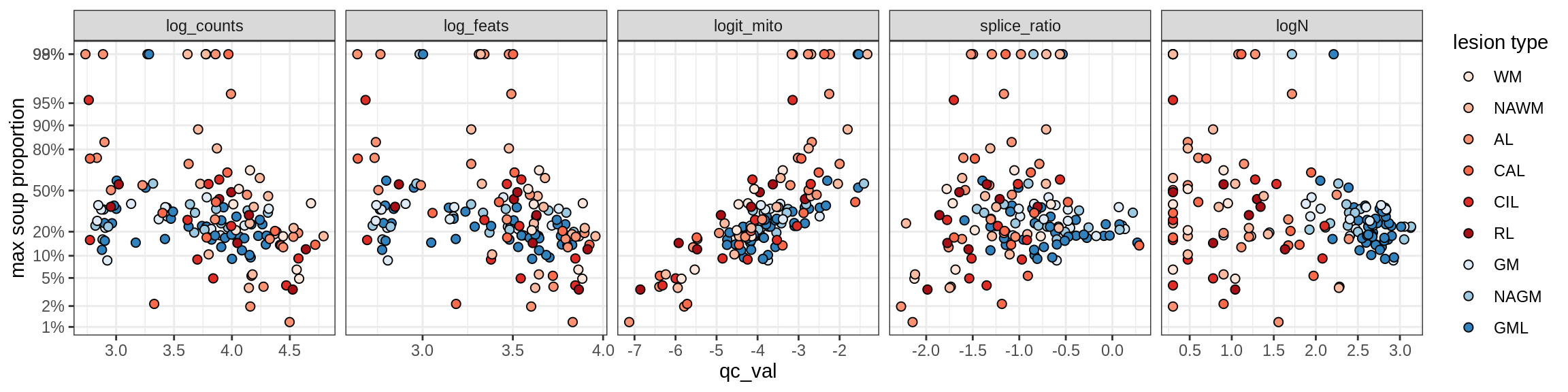
Ex_RORB_A
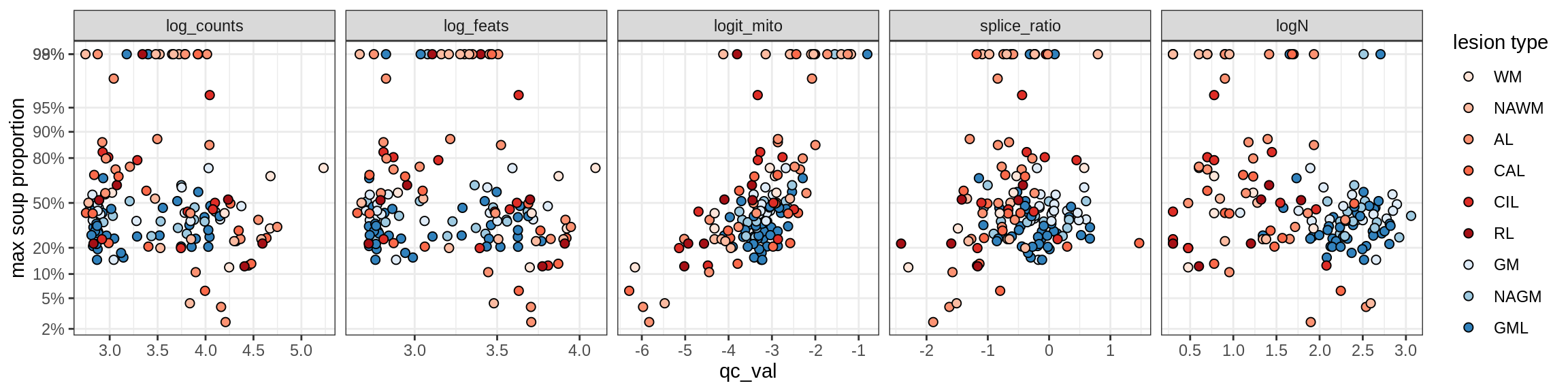
Ex_RORB_B
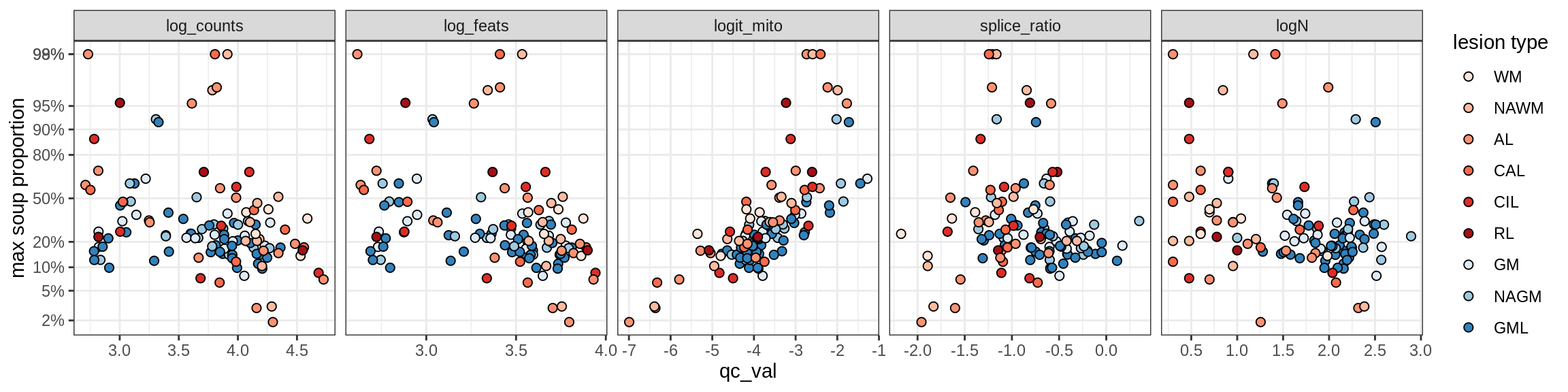
Ex_RORB_C
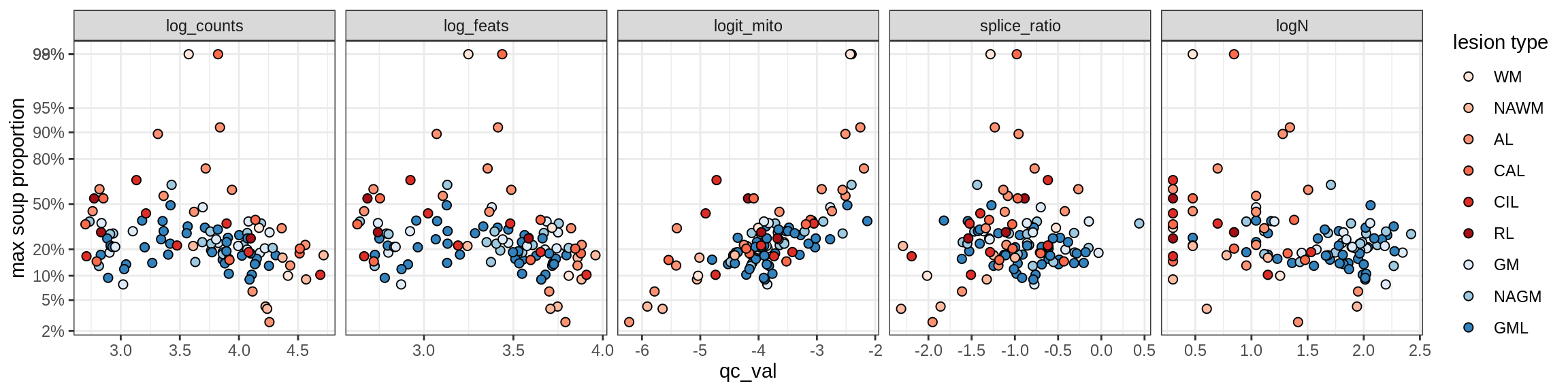
Ex_RORB_CUX2_A
Ex_RORB_CUX2_B
Ex_RORB_CUX2_C
Ex_RORB_CUX2_D
Ex_RORB_CUX2_E
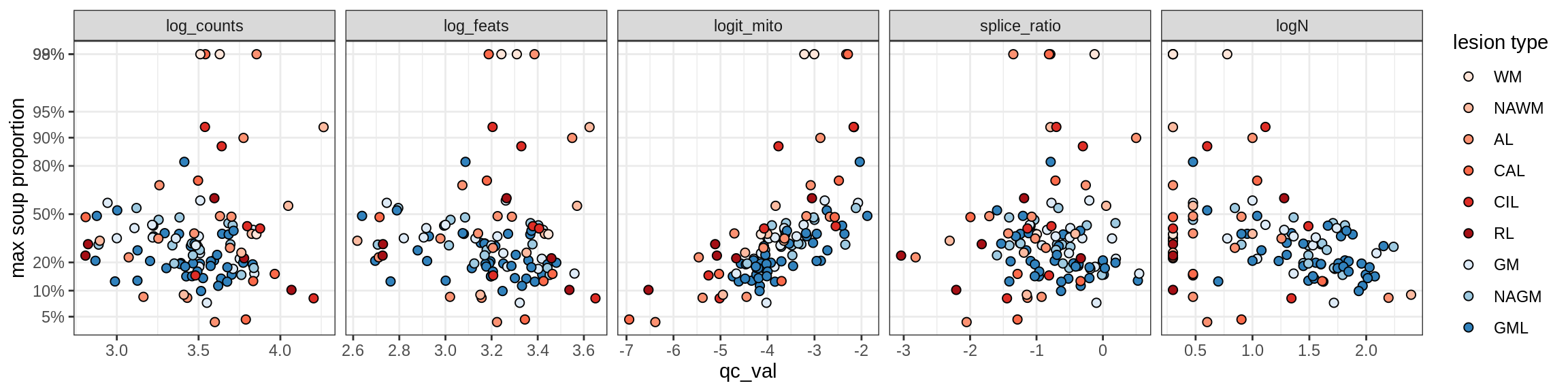
Ex_RORB_CUX2_F
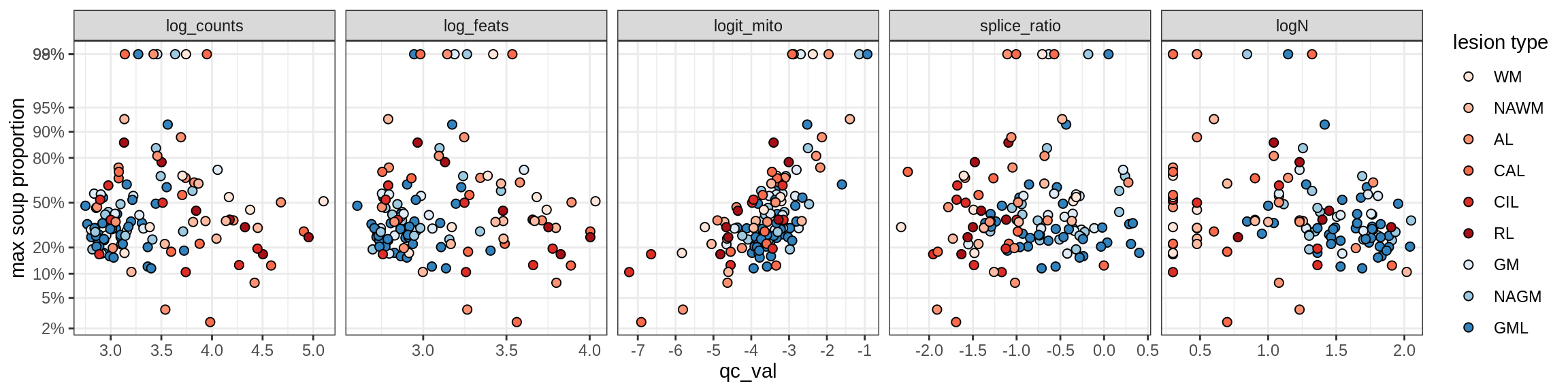
Ex_RORB_THEMIS_A
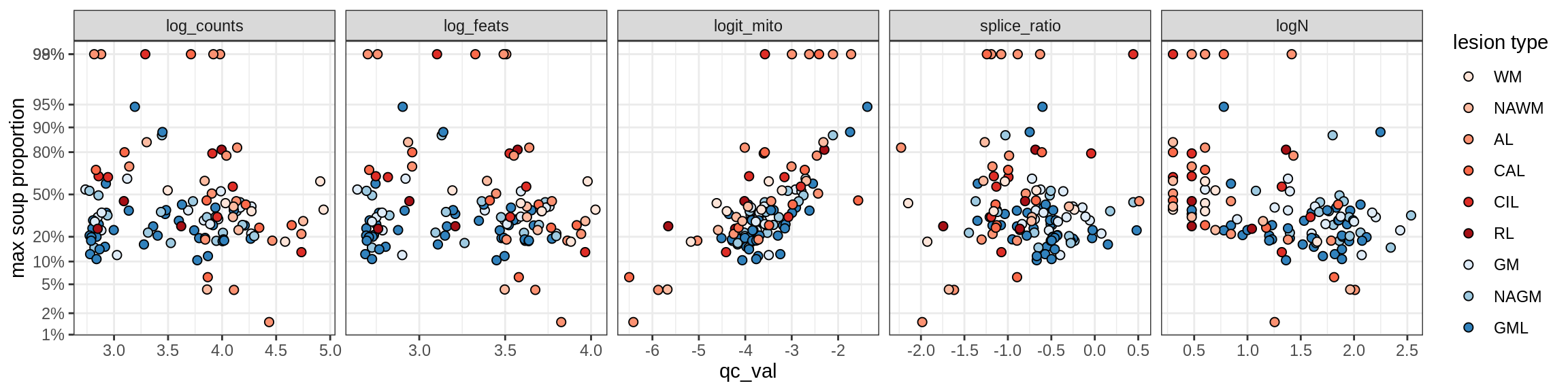
Ex_RORB_THEMIS_B
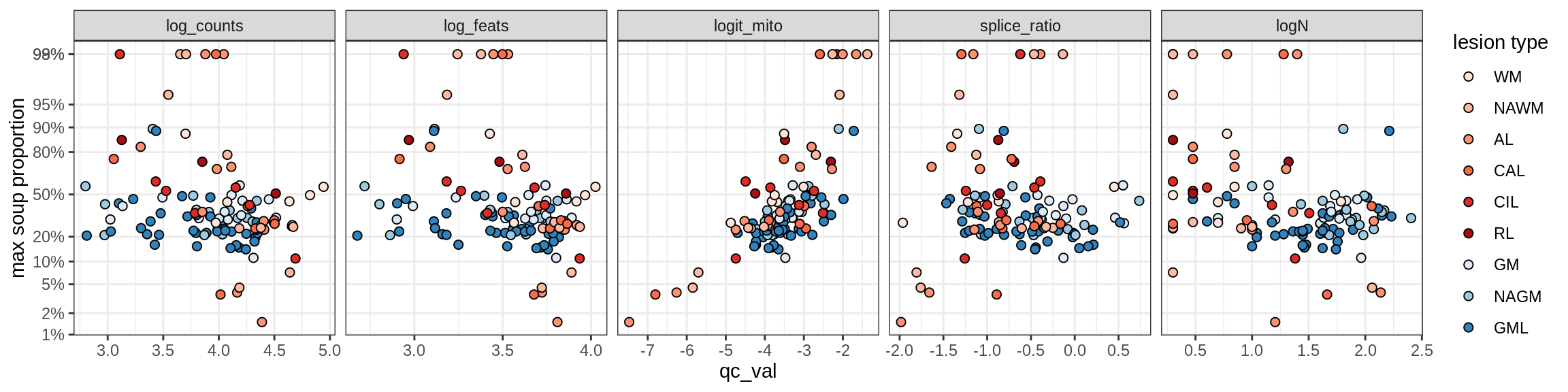
Ex_THEMIS_A
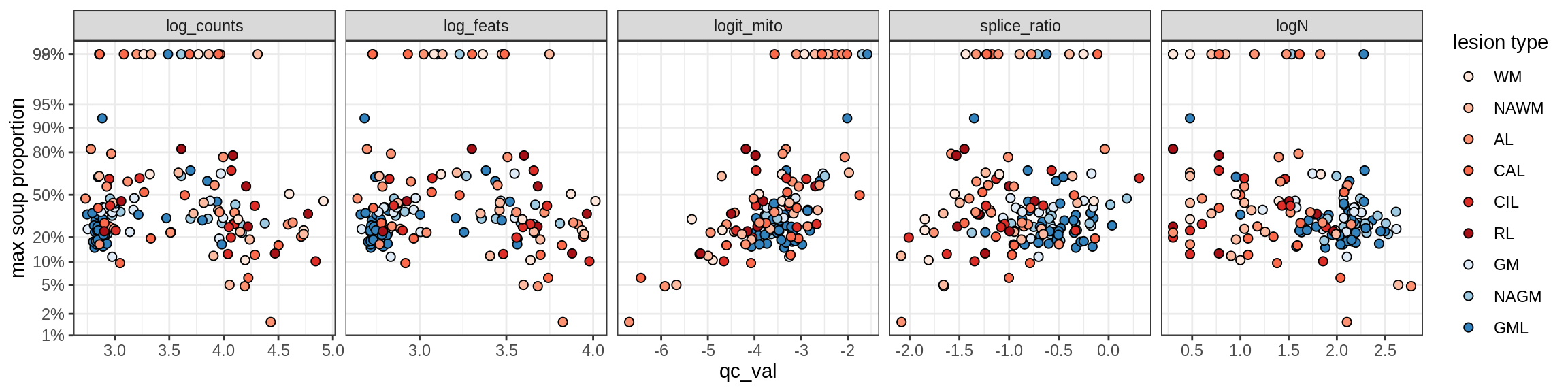
Ex_THEMIS_B
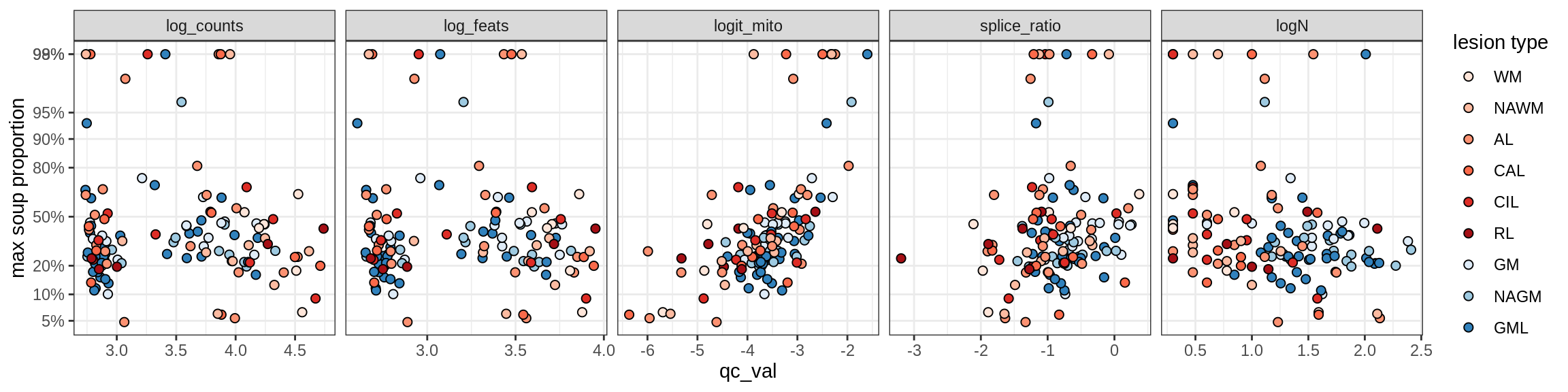
Ex_TLE4_A
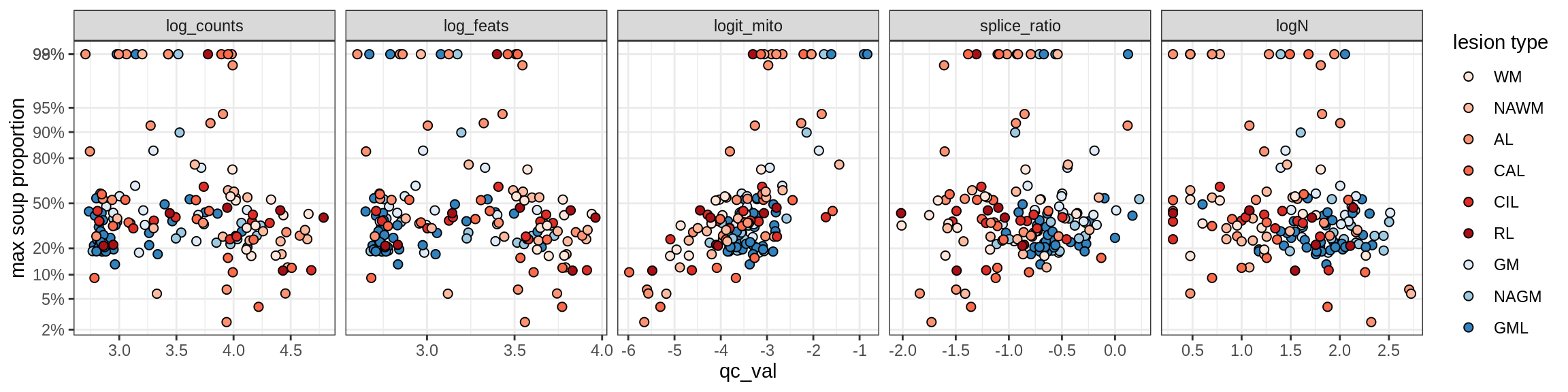
Ex_TLE4_B
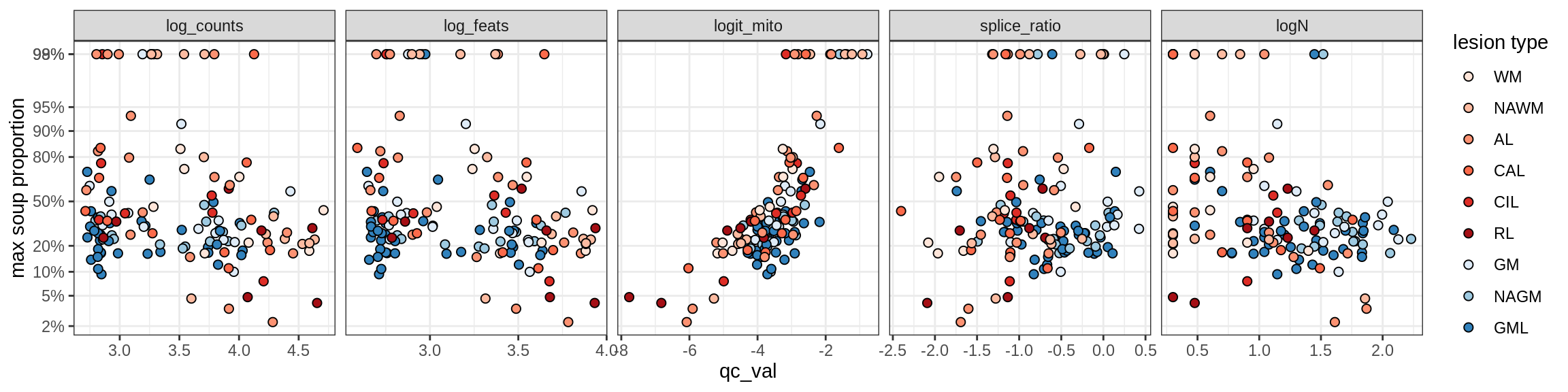
Neuro_oligo
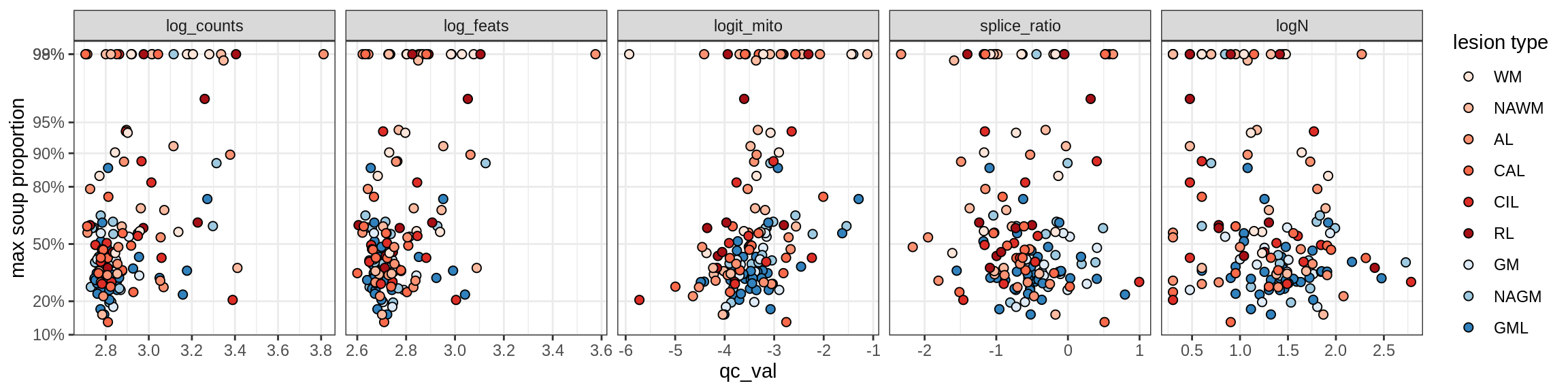
Inhibitory neurons
Inh_LAMP5
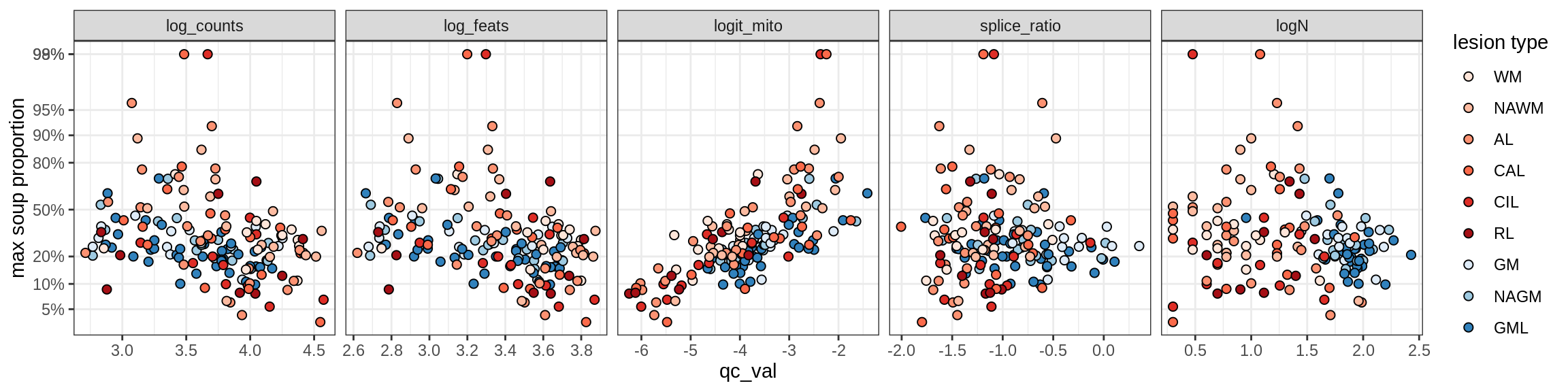
Inh_Pvalb_A
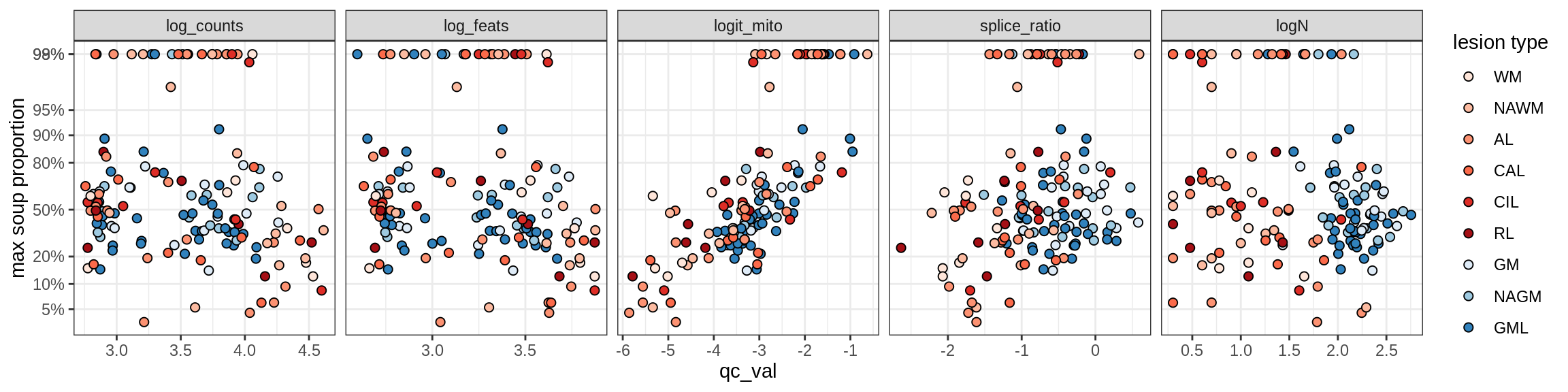
Inh_Pvalb_B
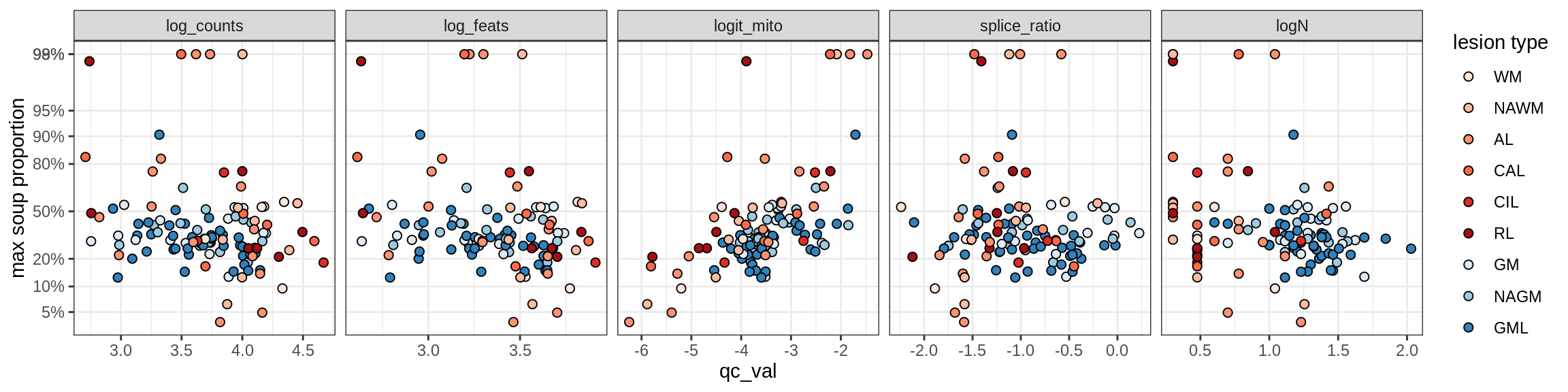
Inh_RELN
Inh_SST_A
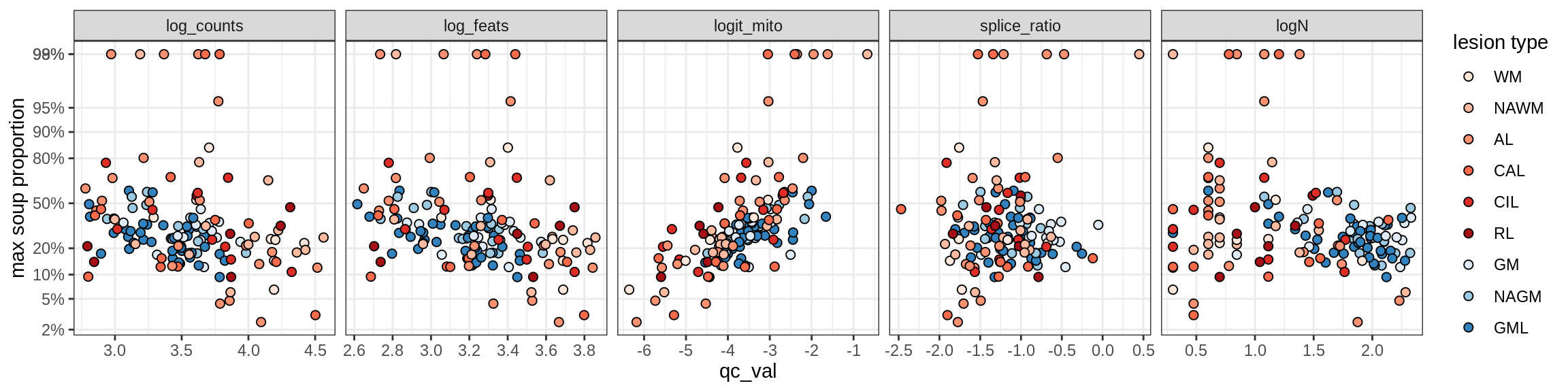
Inh_SST_B
Inh_VIP
Endothelial cells
Endo_A
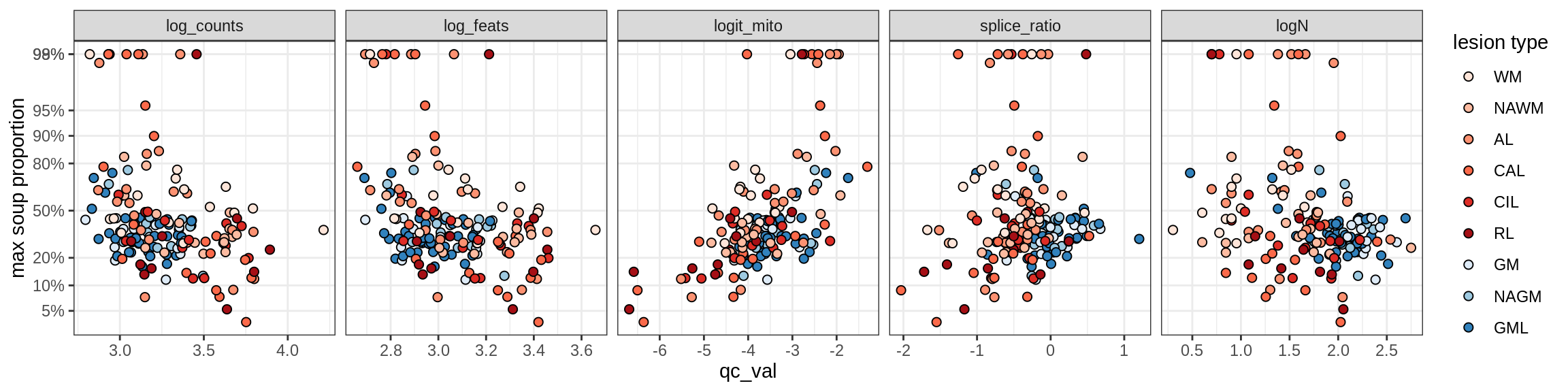
Endo_B
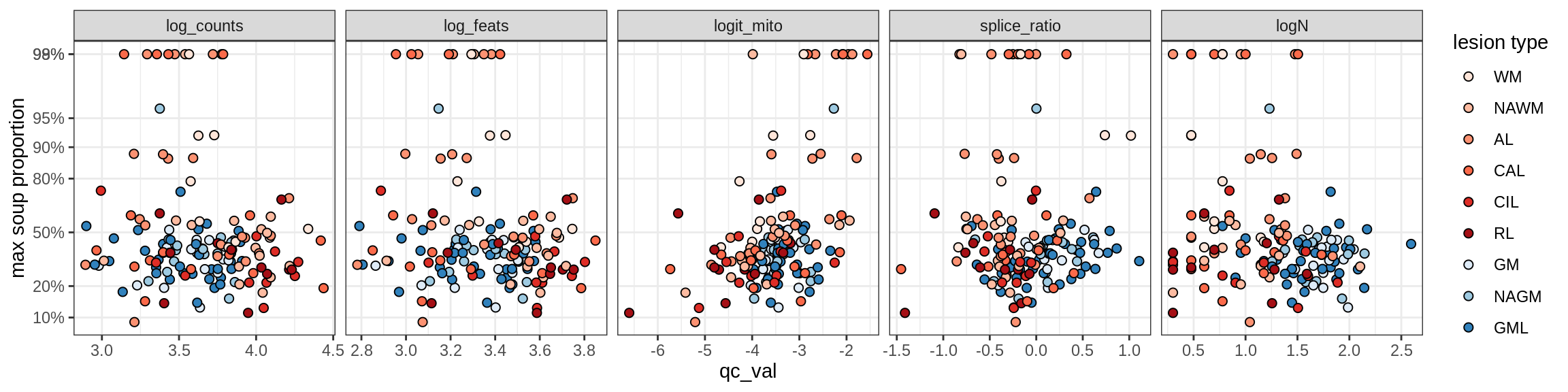
Pericytes
Pericytes
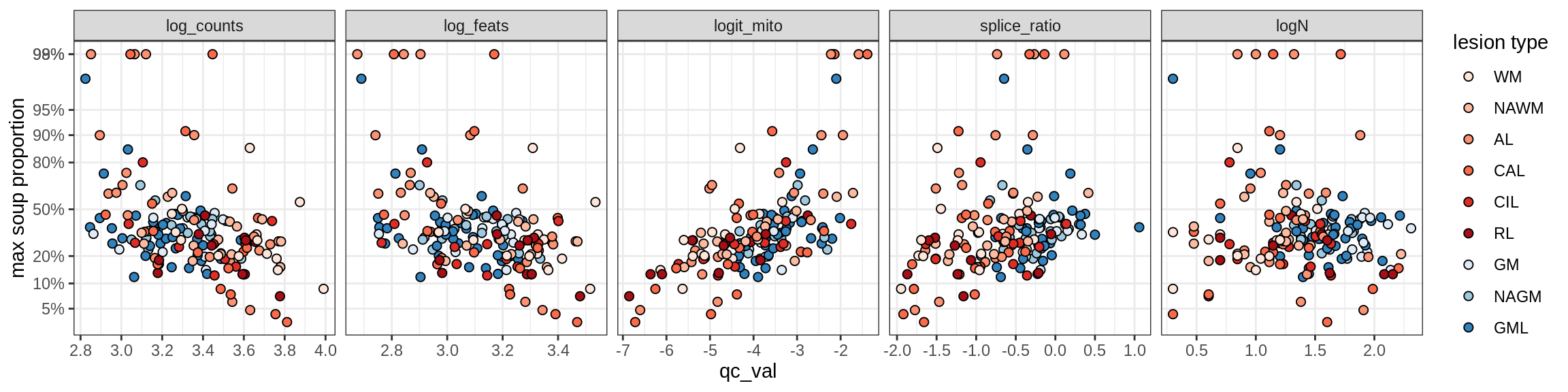
Immune
B_cells
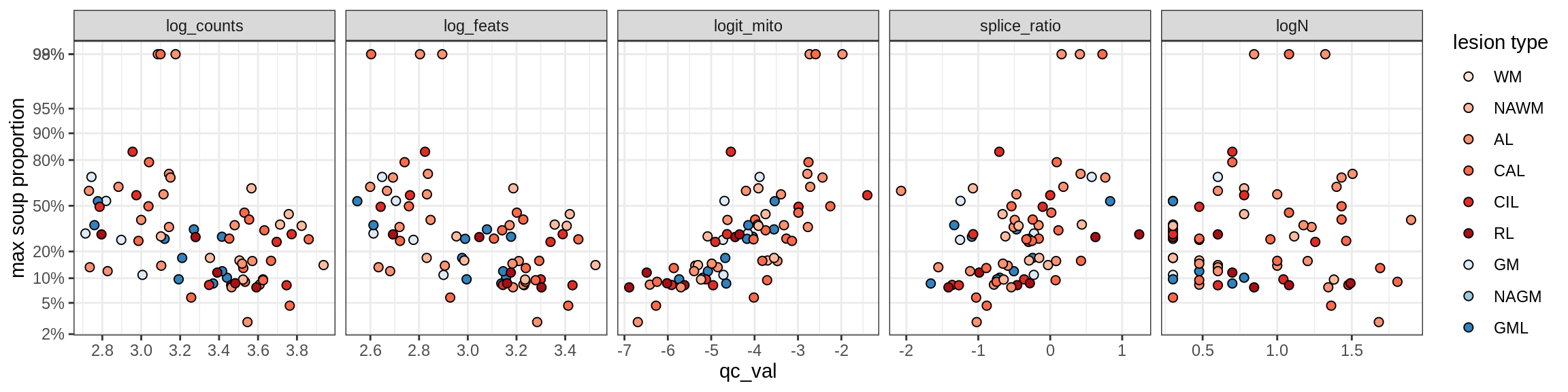
T_cells
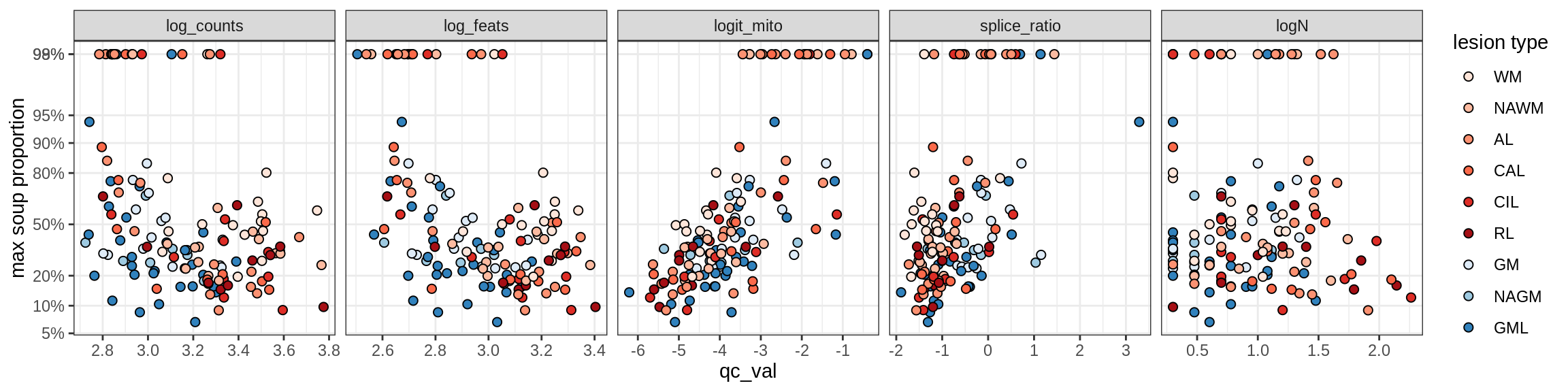
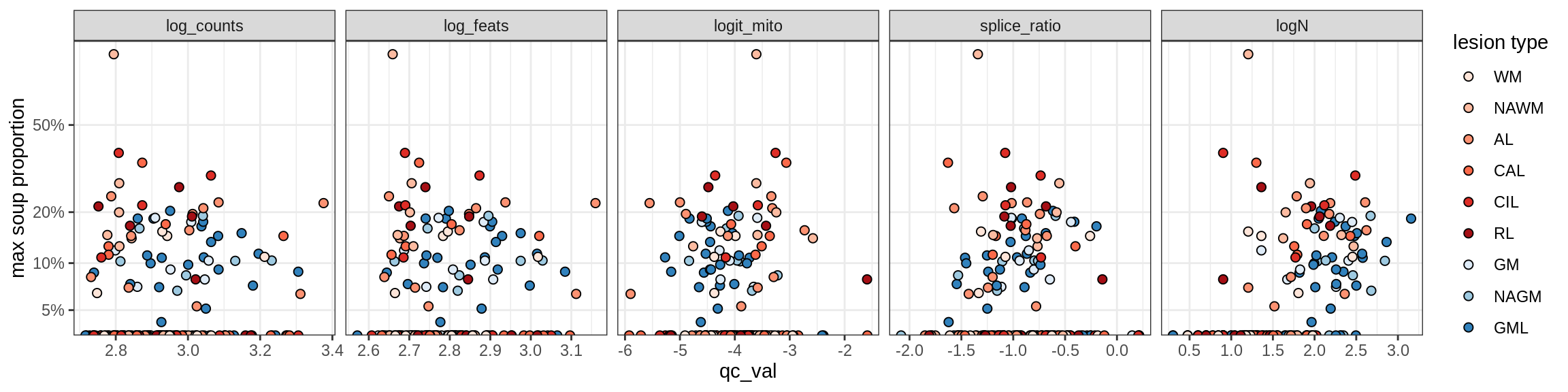
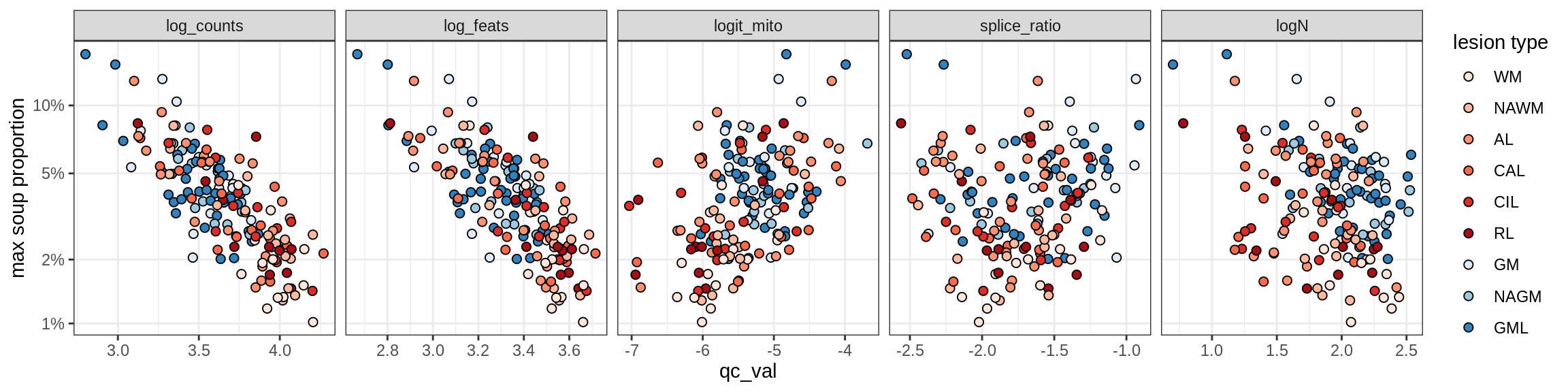
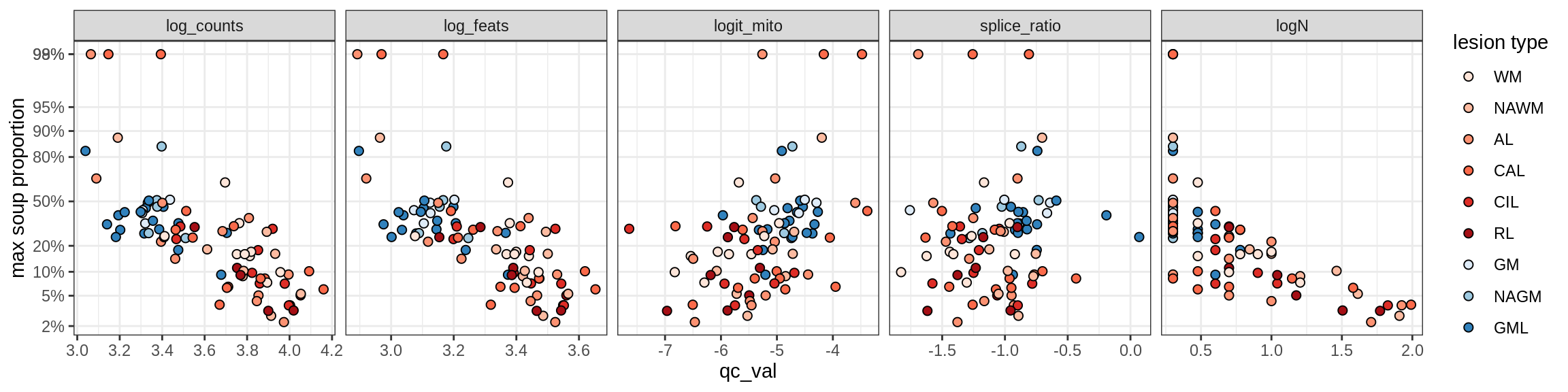
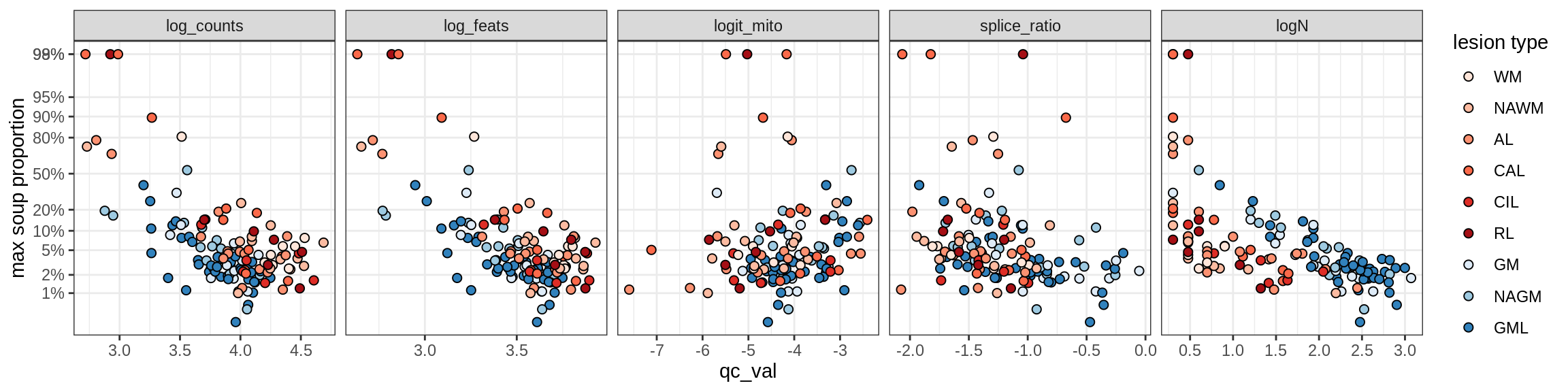
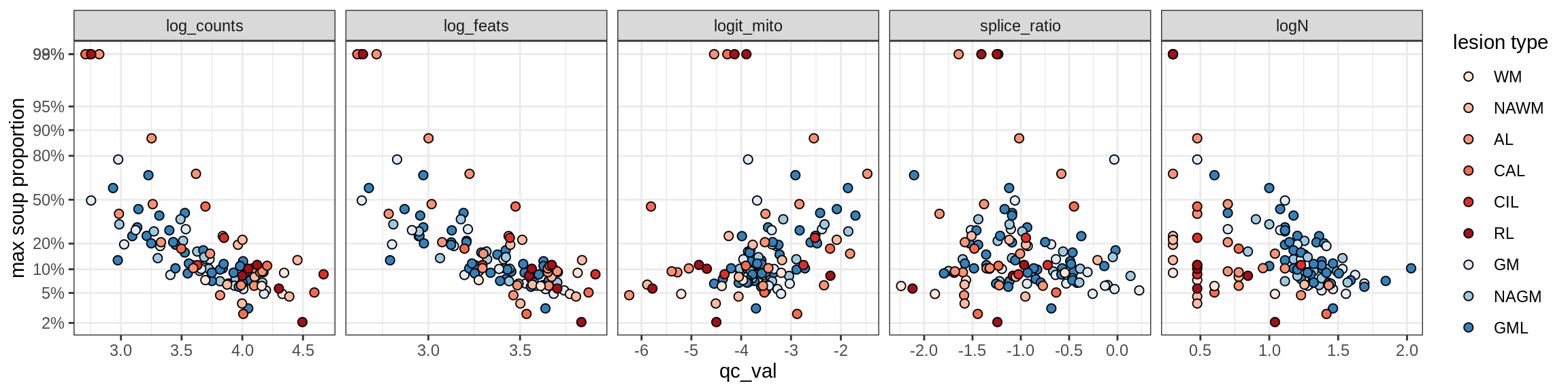
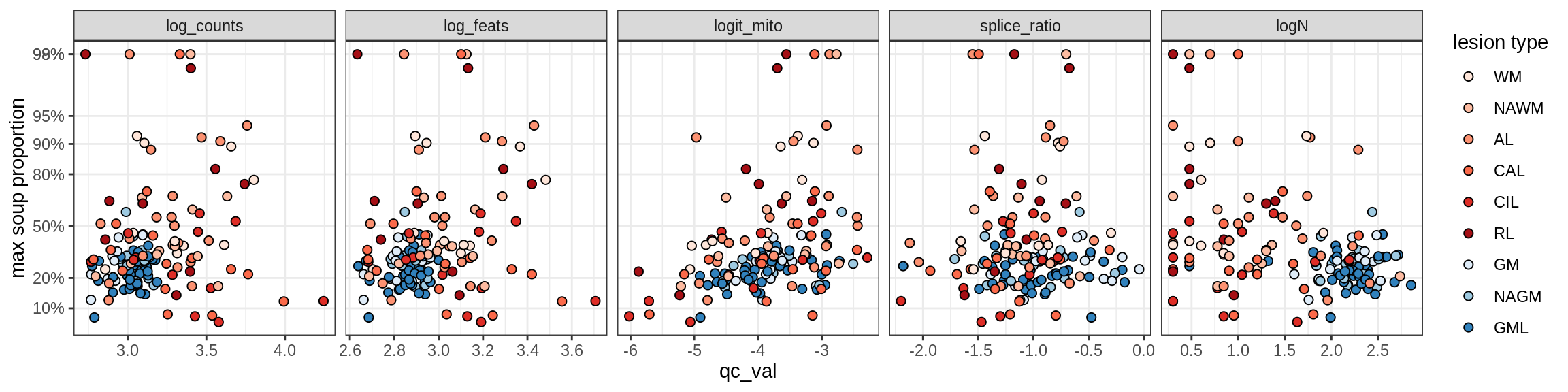
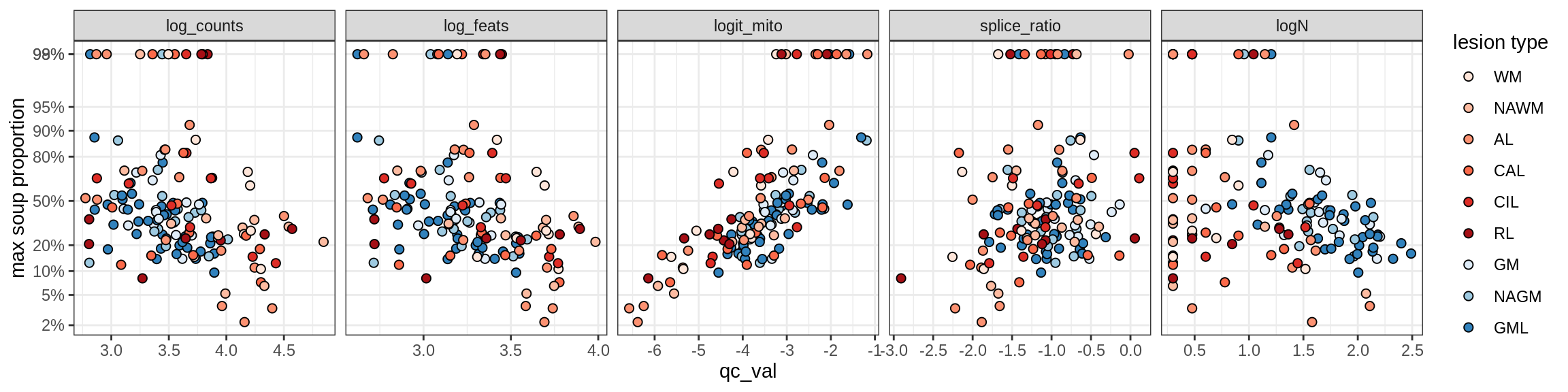
Correlations of “maximum” contamination with QC metrics (comparison of celltypes)
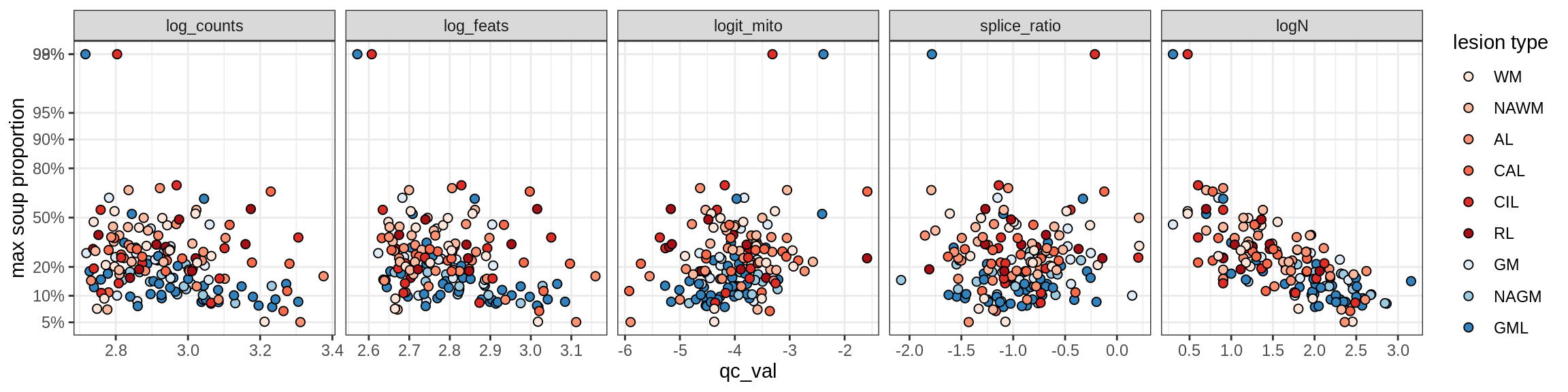
cat('### Random soup (-ve control)\n')Random soup (-ve control)
print(plot_soup_props_vs_qc_by_celltype(props_rand, qc_stats))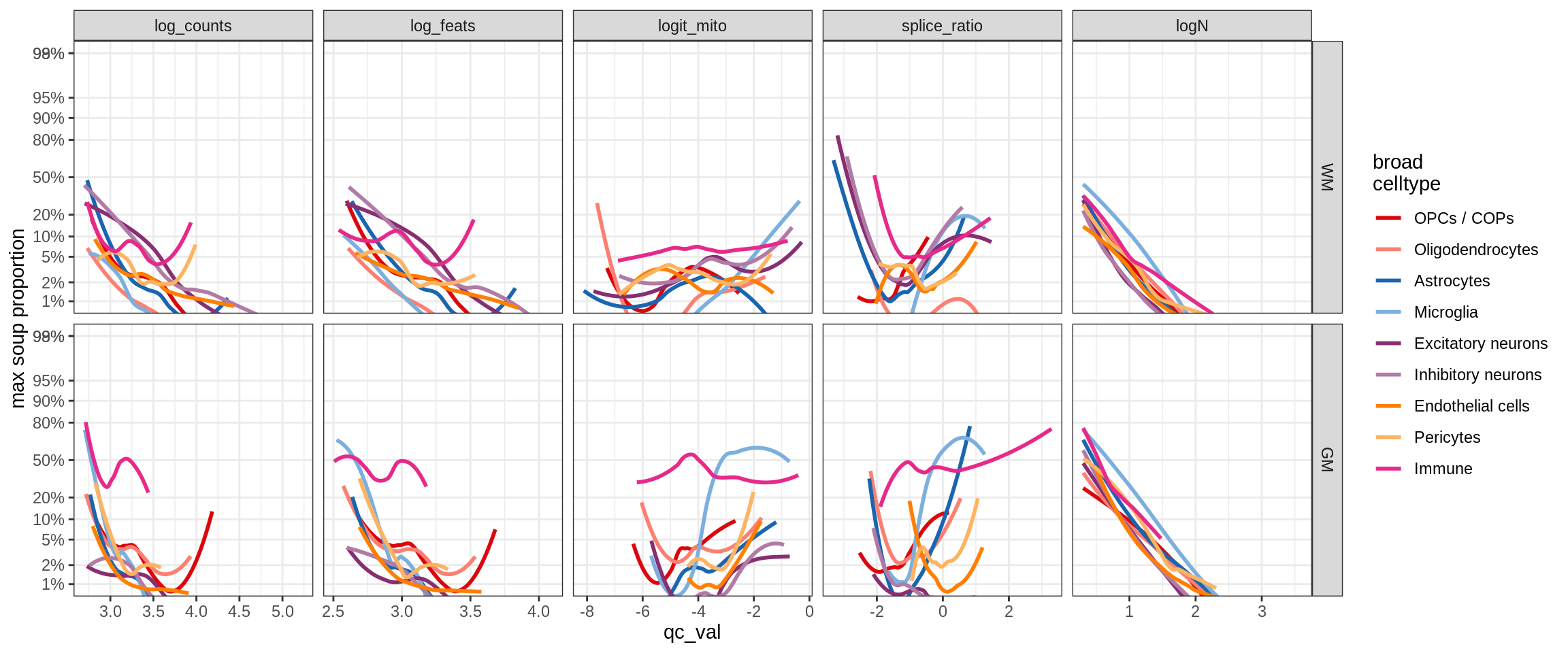
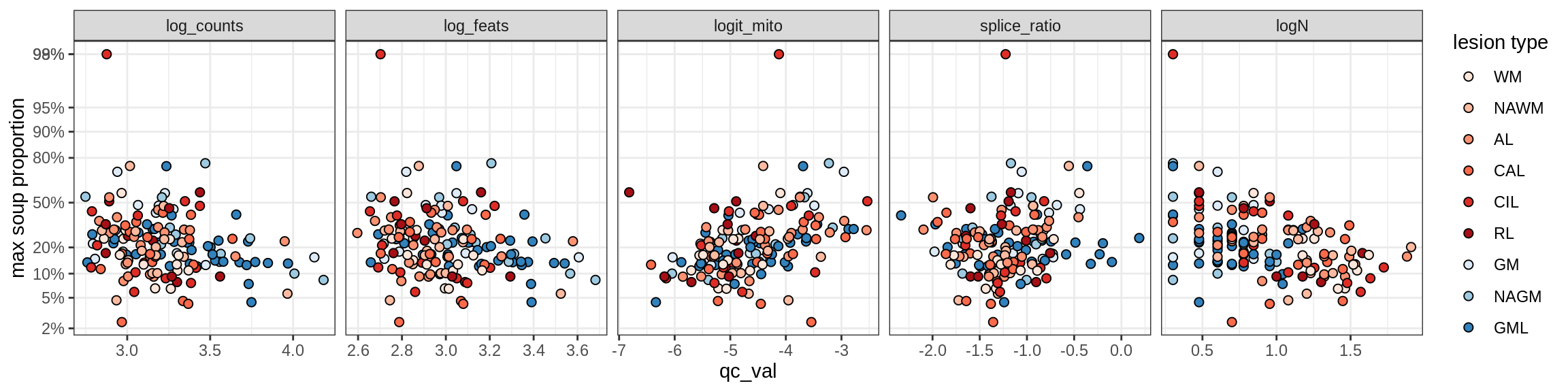
cat('\n\n')cat('### All samples\n')All samples
print(plot_soup_props_vs_qc_by_celltype(props_max, qc_stats))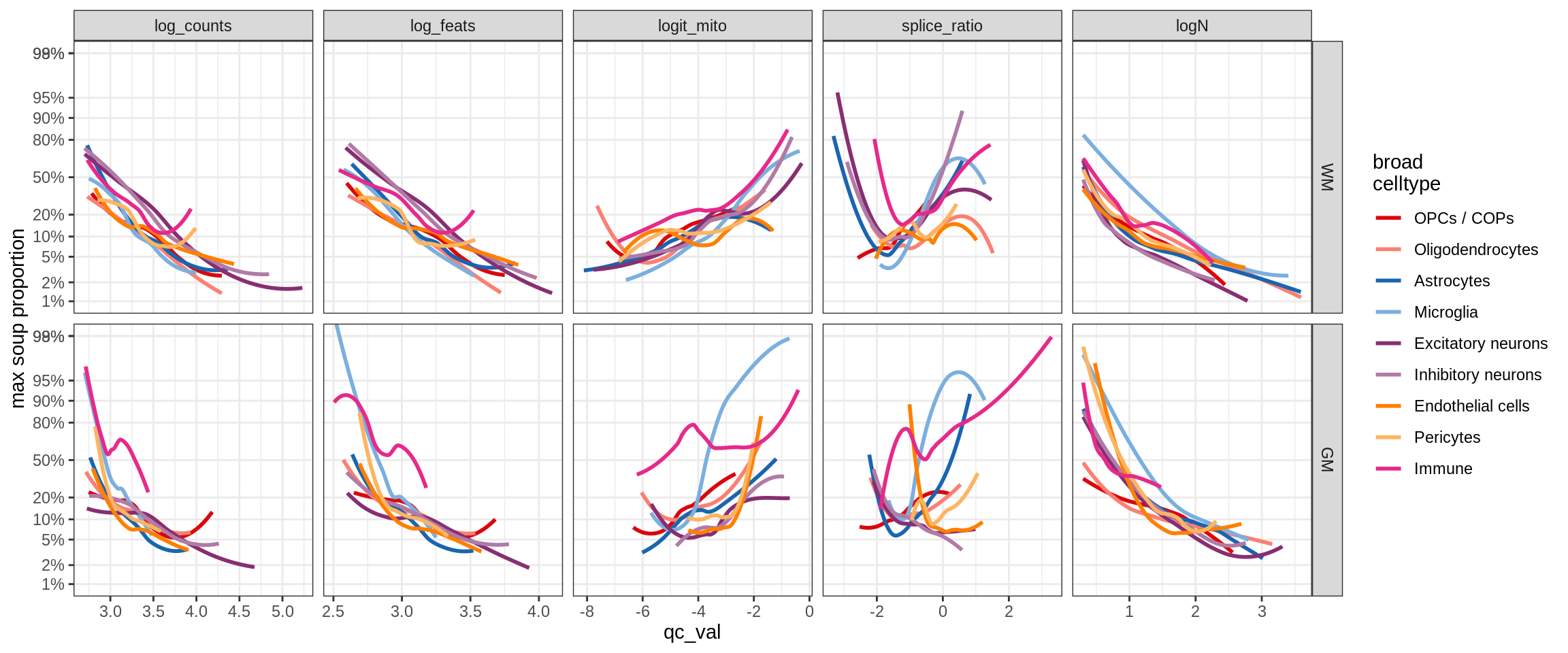
cat('\n\n')cat('### >= 10 cells in pseudobulk\n')>= 10 cells in pseudobulk
big_cls = qc_stats[qc_var == 'logN' & qc_val >= log10(10), .(sample_id, conos)]
stats_big = merge(qc_stats, big_cls, by = c('sample_id', 'conos'))
print(plot_soup_props_vs_qc_by_celltype(props_max, stats_big))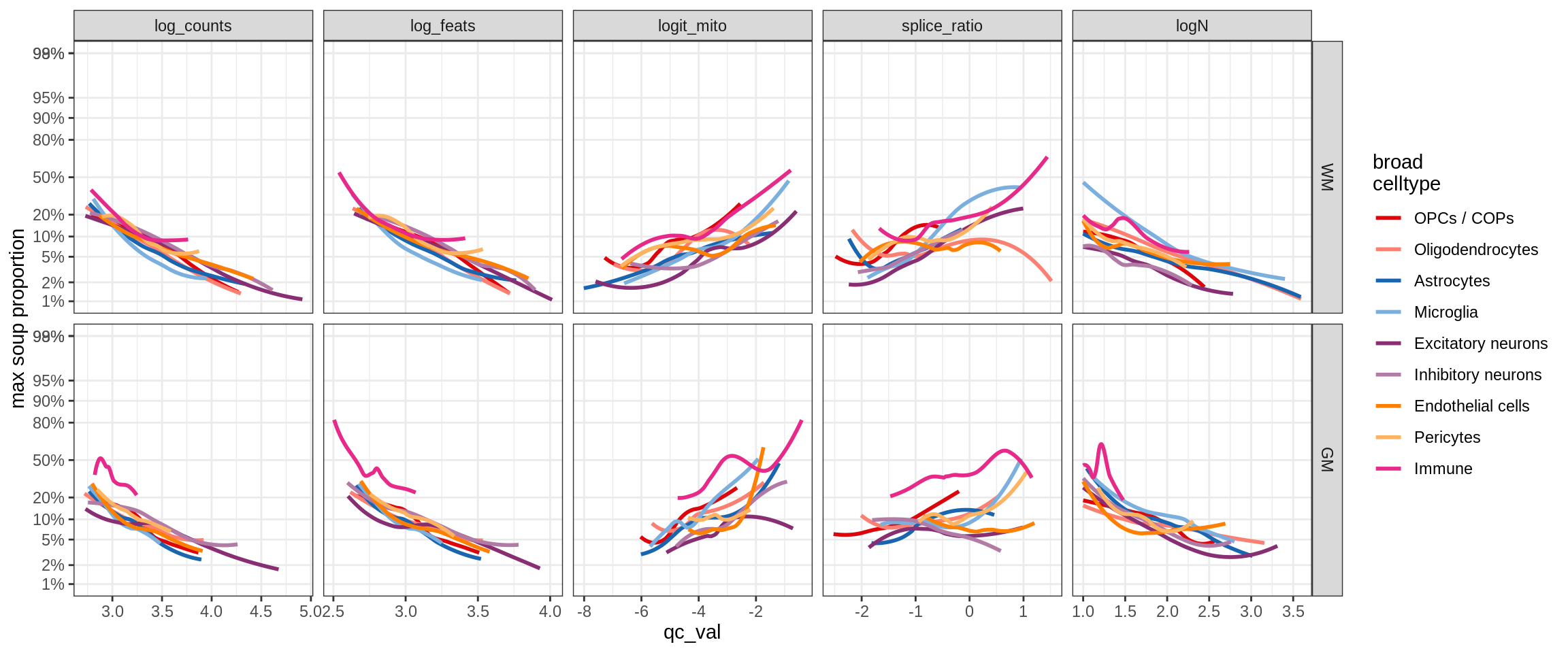
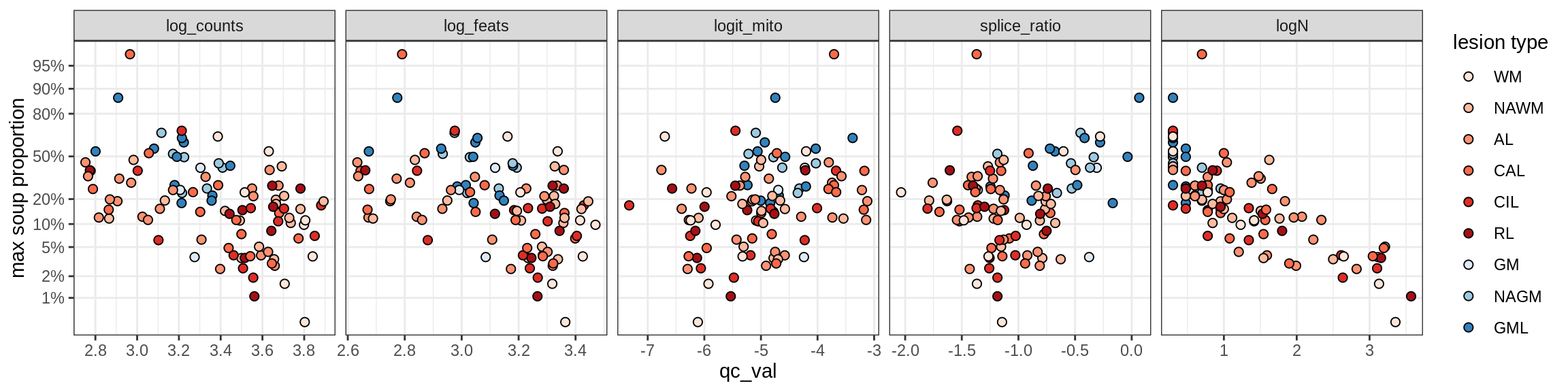
cat('\n\n')Correlations of mito contamination with QC metrics (comparison of celltypes)
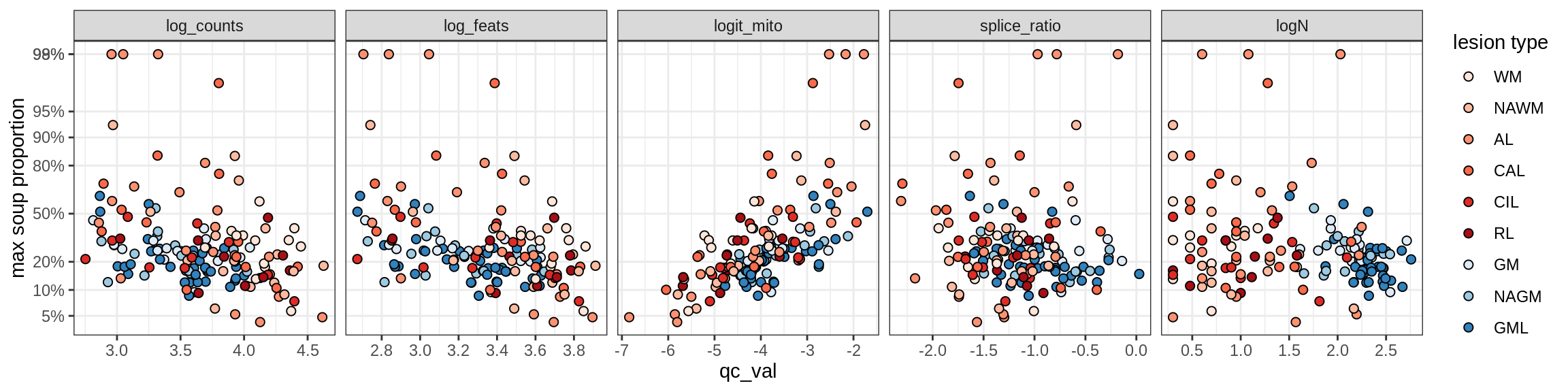
cat('### All samples\n')All samples
print(plot_soup_props_vs_qc_by_celltype(props_mt, qc_stats))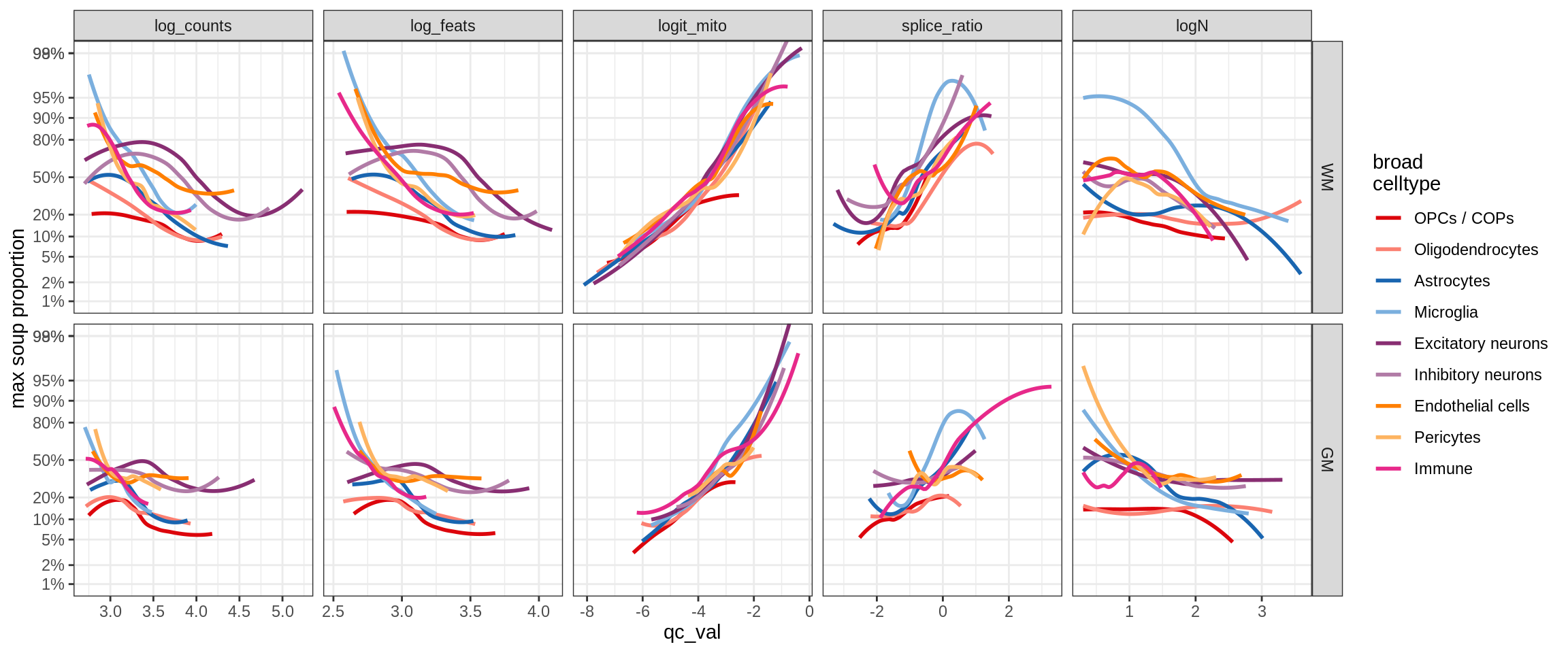
cat('\n\n')cat('### >= 10 cells in pseudobulk\n')>= 10 cells in pseudobulk
big_cls = qc_stats[qc_var == 'logN' & qc_val >= log10(10), .(sample_id, conos)]
stats_big = merge(qc_stats, big_cls, by = c('sample_id', 'conos'))
print(plot_soup_props_vs_qc_by_celltype(props_mt, stats_big))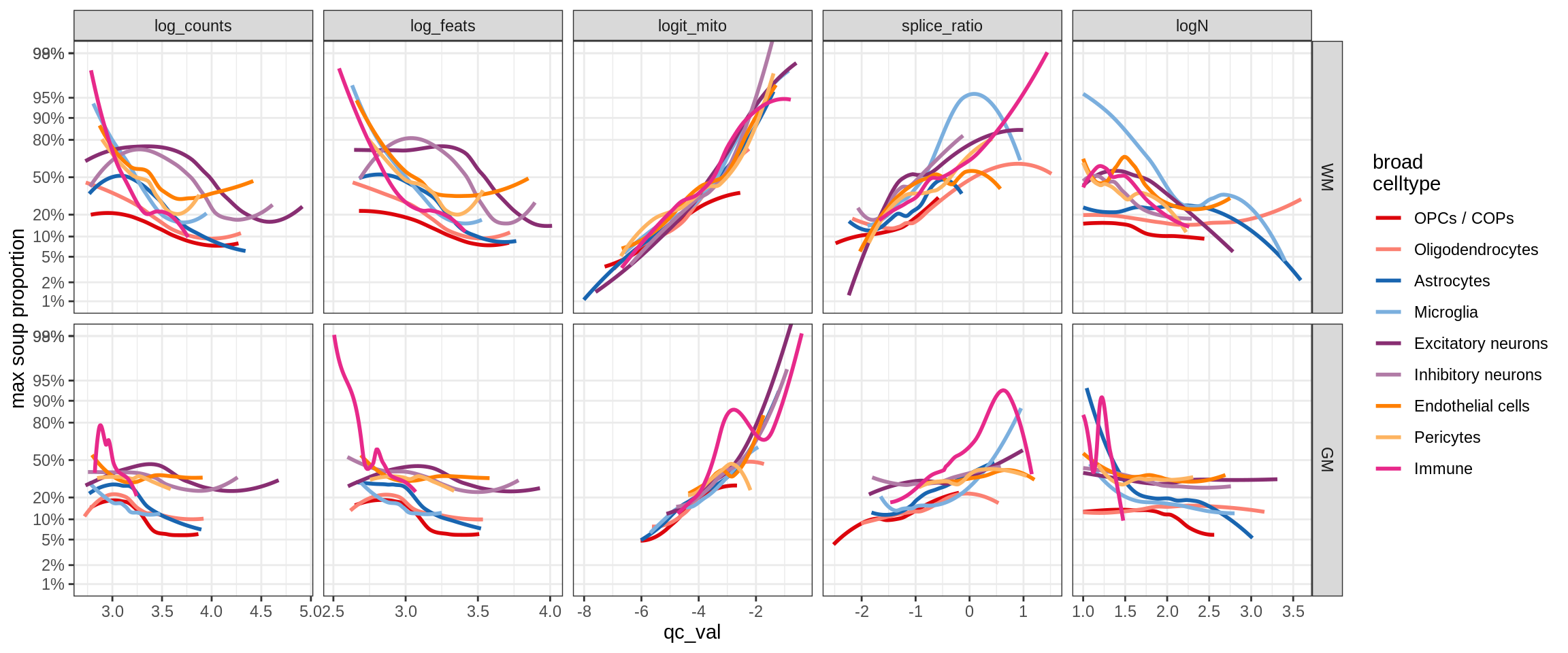
cat('\n\n')Outputs
# save soup proportions, broad
soup_broad = sprintf(soup_q_pat, 'type_broad', 'maximum') %>% readRDS
pb_soup_broad = calc_pb_soup(pb_broad, soup_broad, prof_broad)
saveRDS(pb_soup_broad, file = soup_broad_f, compress = FALSE)
# save soup proportions, fine
soup_fine = sprintf(soup_q_pat, 'type_fine', 'maximum') %>% readRDS
pb_soup_fine = calc_pb_soup(pb_fine, soup_fine, prof_fine)
saveRDS(pb_soup_fine, file = soup_fine_f, compress = FALSE)
# save soup proportions, broad
soup_broad = sprintf(soup_q_pat, 'type_broad', 'mito') %>% readRDS
pb_soup_broad = calc_pb_soup(pb_broad, soup_broad, prof_broad)
saveRDS(pb_soup_broad, file = soup_broad_mito_f, compress = FALSE)
# save soup proportions, fine
soup_fine = sprintf(soup_q_pat, 'type_fine', 'mito') %>% readRDS
pb_soup_fine = calc_pb_soup(pb_fine, soup_fine, prof_fine)
saveRDS(pb_soup_fine, file = soup_fine_mito_f, compress = FALSE)devtools::session_info()Registered S3 method overwritten by 'cli':
method from
print.boxx spatstat.geom- Session info ---------------------------------------------------------------
setting value
version R version 4.0.5 (2021-03-31)
os CentOS Linux 7 (Core)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_US.UTF-8
ctype C
tz Europe/Zurich
date 2021-10-26
- Packages -------------------------------------------------------------------
! package * version date lib
abind 1.4-5 2016-07-21 [2]
annotate 1.68.0 2020-10-27 [1]
AnnotationDbi 1.52.0 2020-10-27 [1]
assertthat * 0.2.1 2019-03-21 [2]
backports 1.2.1 2020-12-09 [2]
beachmat 2.6.4 2020-12-20 [1]
beeswarm 0.4.0 2021-06-01 [1]
Biobase * 2.50.0 2020-10-27 [1]
BiocGenerics * 0.36.1 2021-04-16 [1]
BiocManager 1.30.16 2021-06-15 [1]
BiocNeighbors 1.8.2 2020-12-07 [1]
BiocParallel * 1.24.1 2020-11-06 [1]
BiocSingular 1.6.0 2020-10-27 [1]
BiocStyle * 2.18.1 2020-11-24 [1]
Biostrings 2.58.0 2020-10-27 [1]
bit 4.0.4 2020-08-04 [2]
bit64 4.0.5 2020-08-30 [2]
bitops 1.0-7 2021-04-24 [2]
blme 1.0-5 2021-01-05 [1]
blob 1.2.2 2021-07-23 [2]
bluster 1.0.0 2020-10-27 [1]
boot 1.3-28 2021-05-03 [2]
broom 0.7.9 2021-07-27 [2]
bslib 0.3.1 2021-10-06 [2]
cachem 1.0.6 2021-08-19 [1]
Cairo 1.5-12.2 2020-07-07 [2]
callr 3.7.0 2021-04-20 [2]
caTools 1.18.2 2021-03-28 [2]
cellranger 1.1.0 2016-07-27 [2]
circlize * 0.4.13 2021-06-09 [1]
cli 3.0.1 2021-07-17 [1]
clue 0.3-60 2021-10-11 [1]
cluster 2.1.2 2021-04-17 [2]
codetools 0.2-18 2020-11-04 [2]
colorout * 1.2-2 2021-04-15 [1]
colorRamps 2.3 2012-10-29 [1]
colorspace 2.0-2 2021-06-24 [1]
ComplexHeatmap * 2.6.2 2020-11-12 [1]
conos * 1.4.3 2021-08-07 [1]
cowplot 1.1.1 2020-12-30 [2]
crayon 1.4.1 2021-02-08 [2]
data.table * 1.14.2 2021-09-27 [2]
DBI 1.1.1 2021-01-15 [2]
DelayedArray 0.16.3 2021-03-24 [1]
DelayedMatrixStats 1.12.3 2021-02-03 [1]
deldir 1.0-6 2021-10-23 [2]
desc 1.4.0 2021-09-28 [1]
DESeq2 1.30.1 2021-02-19 [1]
devtools 2.4.2 2021-06-07 [1]
digest 0.6.28 2021-09-23 [2]
doParallel 1.0.16 2020-10-16 [1]
dplyr 1.0.7 2021-06-18 [2]
dqrng 0.3.0 2021-05-01 [2]
DropletUtils * 1.10.3 2021-02-02 [1]
edgeR * 3.32.1 2021-01-14 [1]
ellipsis 0.3.2 2021-04-29 [2]
evaluate 0.14 2019-05-28 [2]
fansi 0.5.0 2021-05-25 [2]
farver 2.1.0 2021-02-28 [2]
fastmap 1.1.0 2021-01-25 [2]
fitdistrplus 1.1-6 2021-09-28 [2]
forcats * 0.5.1 2021-01-27 [2]
foreach 1.5.1 2020-10-15 [2]
fs 1.5.0 2020-07-31 [2]
future 1.22.1 2021-08-25 [2]
future.apply 1.8.1 2021-08-10 [2]
genefilter 1.72.1 2021-01-21 [1]
geneplotter 1.68.0 2020-10-27 [1]
generics 0.1.1 2021-10-25 [2]
GenomeInfoDb * 1.26.7 2021-04-08 [1]
GenomeInfoDbData 1.2.4 2021-04-15 [1]
GenomicAlignments 1.26.0 2020-10-27 [1]
GenomicRanges * 1.42.0 2020-10-27 [1]
GetoptLong 1.0.5 2020-12-15 [1]
ggbeeswarm 0.6.0 2017-08-07 [1]
ggplot.multistats * 1.0.0 2019-10-28 [1]
ggplot2 * 3.3.5 2021-06-25 [1]
ggrepel 0.9.1 2021-01-15 [2]
ggridges 0.5.3 2021-01-08 [2]
git2r 0.28.0 2021-01-10 [1]
glmmTMB 1.1.2.3 2021-09-20 [1]
GlobalOptions 0.1.2 2020-06-10 [1]
globals 0.14.0 2020-11-22 [2]
glue 1.4.2 2020-08-27 [2]
goftest 1.2-3 2021-10-07 [2]
googlesheets * 0.3.0 2018-06-29 [1]
gplots 3.1.1 2020-11-28 [2]
gridExtra 2.3 2017-09-09 [2]
grr 0.9.5 2016-08-26 [1]
gtable 0.3.0 2019-03-25 [2]
gtools 3.9.2 2021-06-06 [2]
HDF5Array 1.18.1 2021-02-04 [1]
hdf5r * 1.3.3 2020-08-18 [2]
hexbin 1.28.2 2021-01-08 [2]
highr 0.9 2021-04-16 [2]
hms 1.1.1 2021-09-26 [1]
htmltools 0.5.2 2021-08-25 [2]
htmlwidgets 1.5.4 2021-09-08 [2]
httpuv 1.6.3 2021-09-09 [2]
httr 1.4.2 2020-07-20 [2]
ica 1.0-2 2018-05-24 [2]
igraph * 1.2.7 2021-10-15 [2]
IRanges * 2.24.1 2020-12-12 [1]
irlba 2.3.3 2019-02-05 [2]
iterators * 1.0.13 2020-10-15 [2]
itertools * 0.1-3 2014-03-12 [1]
janitor 2.1.0 2021-01-05 [1]
jquerylib 0.1.4 2021-04-26 [2]
jsonlite 1.7.2 2020-12-09 [2]
kernlab 0.9-29 2019-11-12 [1]
KernSmooth 2.23-20 2021-05-03 [2]
knitr 1.36 2021-09-29 [1]
labeling 0.4.2 2020-10-20 [2]
later 1.3.0 2021-08-18 [2]
lattice 0.20-45 2021-09-22 [2]
lazyeval 0.2.2 2019-03-15 [2]
R leiden 0.3.8 <NA> [2]
leidenAlg 0.1.1 2021-03-03 [1]
lifecycle 1.0.1 2021-09-24 [2]
limma * 3.46.0 2020-10-27 [1]
listenv 0.8.0 2019-12-05 [2]
lme4 1.1-27.1 2021-06-22 [1]
lmerTest 3.1-3 2020-10-23 [1]
lmtest 0.9-38 2020-09-09 [2]
locfit 1.5-9.4 2020-03-25 [1]
loomR * 0.2.0 2021-04-15 [1]
lubridate 1.8.0 2021-10-07 [2]
magrittr * 2.0.1 2020-11-17 [1]
MASS 7.3-54 2021-05-03 [2]
Matrix * 1.3-4 2021-06-01 [2]
Matrix.utils 0.9.8 2020-02-26 [1]
MatrixGenerics * 1.2.1 2021-01-30 [1]
matrixStats * 0.61.0 2021-09-17 [1]
mclust 5.4.7 2020-11-20 [1]
memoise 2.0.0 2021-01-26 [1]
mgcv 1.8-38 2021-10-06 [1]
mime 0.12 2021-09-28 [1]
miniUI 0.1.1.1 2018-05-18 [2]
minqa 1.2.4 2014-10-09 [1]
mixtools 1.2.0 2020-02-07 [1]
munsell 0.5.0 2018-06-12 [2]
muscat * 1.5.1 2021-04-15 [1]
mvnfast 0.2.7 2021-05-20 [1]
mvtnorm 1.1-3 2021-10-08 [1]
nlme 3.1-153 2021-09-07 [2]
nloptr 1.2.2.2 2020-07-02 [1]
nnls * 1.4 2012-03-19 [1]
numDeriv 2016.8-1.1 2019-06-06 [2]
parallelly 1.28.1 2021-09-09 [2]
patchwork * 1.1.1 2020-12-17 [2]
pbapply 1.5-0 2021-09-16 [2]
pbkrtest 0.5.1 2021-03-09 [1]
pillar 1.6.4 2021-10-18 [1]
pkgbuild 1.2.0 2020-12-15 [1]
pkgconfig 2.0.3 2019-09-22 [2]
pkgload 1.2.3 2021-10-13 [2]
plotly 4.10.0 2021-10-09 [2]
plyr 1.8.6 2020-03-03 [2]
png 0.1-7 2013-12-03 [2]
polyclip 1.10-0 2019-03-14 [2]
prettyunits 1.1.1 2020-01-24 [2]
processx 3.5.2 2021-04-30 [2]
progress 1.2.2 2019-05-16 [2]
promises 1.2.0.1 2021-02-11 [2]
ps 1.6.0 2021-02-28 [2]
purrr 0.3.4 2020-04-17 [2]
R.methodsS3 1.8.1 2020-08-26 [1]
R.oo 1.24.0 2020-08-26 [1]
R.utils 2.11.0 2021-09-26 [1]
R6 * 2.5.1 2021-08-19 [2]
RANN 2.6.1 2019-01-08 [2]
RColorBrewer * 1.1-2 2014-12-07 [2]
Rcpp 1.0.7 2021-07-07 [1]
RcppAnnoy 0.0.19 2021-07-30 [1]
RCurl 1.98-1.5 2021-09-17 [1]
readxl * 1.3.1 2019-03-13 [2]
registry 0.5-1 2019-03-05 [1]
remotes 2.4.1 2021-09-29 [1]
reshape2 1.4.4 2020-04-09 [2]
reticulate 1.22 2021-09-17 [2]
rhdf5 2.34.0 2020-10-27 [1]
rhdf5filters 1.2.1 2021-05-03 [1]
Rhdf5lib 1.12.1 2021-01-26 [1]
rjson 0.2.20 2018-06-08 [1]
rlang 0.4.12 2021-10-18 [2]
rmarkdown 2.11 2021-09-14 [1]
ROCR 1.0-11 2020-05-02 [2]
rpart 4.1-15 2019-04-12 [2]
rprojroot 2.0.2 2020-11-15 [2]
Rsamtools 2.6.0 2020-10-27 [1]
RSQLite 2.2.8 2021-08-21 [1]
rsvd 1.0.5 2021-04-16 [1]
rtracklayer 1.50.0 2020-10-27 [1]
Rtsne 0.15 2018-11-10 [2]
S4Vectors * 0.28.1 2020-12-09 [1]
SampleQC * 0.6.1 2021-08-16 [1]
sass 0.4.0 2021-05-12 [2]
scales * 1.1.1 2020-05-11 [2]
scater * 1.18.6 2021-02-26 [1]
scattermore 0.7 2020-11-24 [2]
sccore 1.0.0 2021-10-07 [1]
scran * 1.18.7 2021-04-16 [1]
sctransform 0.3.2 2020-12-16 [2]
scuttle 1.0.4 2020-12-17 [1]
segmented 1.3-4 2021-04-22 [1]
seriation * 1.3.1 2021-10-16 [1]
sessioninfo 1.1.1 2018-11-05 [1]
Seurat * 4.0.5 2021-10-17 [2]
SeuratObject * 4.0.2 2021-06-09 [2]
shape 1.4.6 2021-05-19 [1]
shiny 1.7.1 2021-10-02 [2]
SingleCellExperiment * 1.12.0 2020-10-27 [1]
snakecase 0.11.0 2019-05-25 [1]
sparseMatrixStats 1.2.1 2021-02-02 [1]
spatstat.core 2.3-0 2021-07-16 [2]
spatstat.data 2.1-0 2021-03-21 [2]
spatstat.geom 2.3-0 2021-10-09 [2]
spatstat.sparse 2.0-0 2021-03-16 [2]
spatstat.utils 2.2-0 2021-06-14 [2]
statmod 1.4.36 2021-05-10 [1]
stringi 1.7.4 2021-08-25 [1]
stringr * 1.4.0 2019-02-10 [2]
SummarizedExperiment * 1.20.0 2020-10-27 [1]
survival 3.2-13 2021-08-24 [2]
tensor 1.5 2012-05-05 [2]
testthat 3.1.0 2021-10-04 [2]
tibble 3.1.5 2021-09-30 [1]
tidyr 1.1.4 2021-09-27 [2]
tidyselect 1.1.1 2021-04-30 [2]
TMB 1.7.22 2021-09-28 [1]
TSP 1.1-11 2021-10-06 [1]
usethis 2.1.2 2021-10-25 [1]
utf8 1.2.2 2021-07-24 [1]
uwot * 0.1.10 2020-12-15 [2]
variancePartition 1.20.0 2020-10-27 [1]
vctrs 0.3.8 2021-04-29 [2]
vipor 0.4.5 2017-03-22 [1]
viridis * 0.6.2 2021-10-13 [1]
viridisLite * 0.4.0 2021-04-13 [1]
whisker 0.4 2019-08-28 [1]
withr 2.4.2 2021-04-18 [2]
workflowr * 1.6.2 2020-04-30 [1]
xfun 0.27 2021-10-18 [1]
XML 3.99-0.8 2021-09-17 [1]
xtable 1.8-4 2019-04-21 [2]
XVector 0.30.0 2020-10-27 [1]
yaml 2.2.1 2020-02-01 [2]
zlibbioc 1.36.0 2020-10-27 [1]
zoo 1.8-9 2021-03-09 [2]
source
CRAN (R 4.0.0)
Bioconductor
Bioconductor
CRAN (R 4.0.0)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
Bioconductor
Bioconductor
CRAN (R 4.0.3)
Bioconductor
Bioconductor
Bioconductor
Bioconductor
Bioconductor
CRAN (R 4.0.2)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Github (jalvesaq/colorout@79931fd)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
Bioconductor
Bioconductor
CRAN (R 4.0.5)
CRAN (R 4.0.5)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.2)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
Bioconductor
Bioconductor
CRAN (R 4.0.5)
Bioconductor
Bioconductor
Bioconductor
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.2)
CRAN (R 4.0.0)
CRAN (R 4.0.5)
Bioconductor
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.0)
<NA>
CRAN (R 4.0.3)
CRAN (R 4.0.5)
Bioconductor
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Github (mojaveazure/loomR@df0144b)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
Github (HelenaLC/muscat@c939663)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.2)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.0)
CRAN (R 4.0.5)
Bioconductor
Bioconductor
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.5)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.0)
Bioconductor
Github (wmacnair/SampleQC@3ae8a08)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.5)
Bioconductor
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.1)
CRAN (R 4.0.5)
Bioconductor
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.5)
CRAN (R 4.0.0)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.1)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.5)
CRAN (R 4.0.5)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
[1] /pstore/home/macnairw/lib/conda_r3.12
[2] /pstore/home/macnairw/.conda/envs/r_4.0.3/lib/R/library
R -- Package was removed from disk.
sessionInfo()R version 4.0.5 (2021-03-31)
Platform: x86_64-conda-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /pstore/home/macnairw/.conda/envs/r_4.0.3/lib/libopenblasp-r0.3.12.so
locale:
[1] LC_CTYPE=C LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] grid parallel stats4 stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] nnls_1.4 muscat_1.5.1
[3] DropletUtils_1.10.3 edgeR_3.32.1
[5] limma_3.46.0 googlesheets_0.3.0
[7] scran_1.18.7 uwot_0.1.10
[9] scater_1.18.6 BiocParallel_1.24.1
[11] ggplot.multistats_1.0.0 seriation_1.3.1
[13] ComplexHeatmap_2.6.2 SeuratObject_4.0.2
[15] Seurat_4.0.5 conos_1.4.3
[17] igraph_1.2.7 SampleQC_0.6.1
[19] SingleCellExperiment_1.12.0 SummarizedExperiment_1.20.0
[21] Biobase_2.50.0 GenomicRanges_1.42.0
[23] GenomeInfoDb_1.26.7 IRanges_2.24.1
[25] S4Vectors_0.28.1 BiocGenerics_0.36.1
[27] MatrixGenerics_1.2.1 matrixStats_0.61.0
[29] Matrix_1.3-4 loomR_0.2.0
[31] itertools_0.1-3 iterators_1.0.13
[33] hdf5r_1.3.3 R6_2.5.1
[35] patchwork_1.1.1 readxl_1.3.1
[37] forcats_0.5.1 ggplot2_3.3.5
[39] scales_1.1.1 viridis_0.6.2
[41] viridisLite_0.4.0 assertthat_0.2.1
[43] stringr_1.4.0 data.table_1.14.2
[45] magrittr_2.0.1 circlize_0.4.13
[47] RColorBrewer_1.1-2 BiocStyle_2.18.1
[49] colorout_1.2-2 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] rsvd_1.0.5 ica_1.0-2
[3] ps_1.6.0 Rsamtools_2.6.0
[5] foreach_1.5.1 lmtest_0.9-38
[7] rprojroot_2.0.2 crayon_1.4.1
[9] spatstat.core_2.3-0 MASS_7.3-54
[11] rhdf5filters_1.2.1 Matrix.utils_0.9.8
[13] nlme_3.1-153 backports_1.2.1
[15] rlang_0.4.12 XVector_0.30.0
[17] ROCR_1.0-11 irlba_2.3.3
[19] callr_3.7.0 nloptr_1.2.2.2
[21] rjson_0.2.20 bit64_4.0.5
[23] glue_1.4.2 sctransform_0.3.2
[25] processx_3.5.2 pbkrtest_0.5.1
[27] vipor_0.4.5 spatstat.sparse_2.0-0
[29] AnnotationDbi_1.52.0 spatstat.geom_2.3-0
[31] tidyselect_1.1.1 usethis_2.1.2
[33] fitdistrplus_1.1-6 variancePartition_1.20.0
[35] XML_3.99-0.8 tidyr_1.1.4
[37] zoo_1.8-9 GenomicAlignments_1.26.0
[39] xtable_1.8-4 evaluate_0.14
[41] cli_3.0.1 scuttle_1.0.4
[43] zlibbioc_1.36.0 miniUI_0.1.1.1
[45] whisker_0.4 bslib_0.3.1
[47] rpart_4.1-15 shiny_1.7.1
[49] BiocSingular_1.6.0 xfun_0.27
[51] clue_0.3-60 pkgbuild_1.2.0
[53] cluster_2.1.2 caTools_1.18.2
[55] TSP_1.1-11 tibble_3.1.5
[57] ggrepel_0.9.1 listenv_0.8.0
[59] Biostrings_2.58.0 png_0.1-7
[61] future_1.22.1 withr_2.4.2
[63] bitops_1.0-7 plyr_1.8.6
[65] cellranger_1.1.0 dqrng_0.3.0
[67] pillar_1.6.4 gplots_3.1.1
[69] GlobalOptions_0.1.2 cachem_1.0.6
[71] fs_1.5.0 kernlab_0.9-29
[73] GetoptLong_1.0.5 DelayedMatrixStats_1.12.3
[75] vctrs_0.3.8 ellipsis_0.3.2
[77] generics_0.1.1 devtools_2.4.2
[79] tools_4.0.5 beeswarm_0.4.0
[81] munsell_0.5.0 DelayedArray_0.16.3
[83] pkgload_1.2.3 fastmap_1.1.0
[85] compiler_4.0.5 abind_1.4-5
[87] httpuv_1.6.3 rtracklayer_1.50.0
[89] segmented_1.3-4 sessioninfo_1.1.1
[91] plotly_4.10.0 GenomeInfoDbData_1.2.4
[93] gridExtra_2.3 glmmTMB_1.1.2.3
[95] lattice_0.20-45 deldir_1.0-6
[97] utf8_1.2.2 later_1.3.0
[99] dplyr_1.0.7 jsonlite_1.7.2
[101] pbapply_1.5-0 sparseMatrixStats_1.2.1
[103] genefilter_1.72.1 lazyeval_0.2.2
[105] promises_1.2.0.1 doParallel_1.0.16
[107] R.utils_2.11.0 goftest_1.2-3
[109] spatstat.utils_2.2-0 reticulate_1.22
[111] rmarkdown_2.11 cowplot_1.1.1
[113] blme_1.0-5 statmod_1.4.36
[115] Rtsne_0.15 HDF5Array_1.18.1
[117] survival_3.2-13 numDeriv_2016.8-1.1
[119] yaml_2.2.1 htmltools_0.5.2
[121] memoise_2.0.0 locfit_1.5-9.4
[123] digest_0.6.28 mime_0.12
[125] registry_0.5-1 RSQLite_2.2.8
[127] future.apply_1.8.1 remotes_2.4.1
[129] blob_1.2.2 R.oo_1.24.0
[131] mvnfast_0.2.7 labeling_0.4.2
[133] splines_4.0.5 Rhdf5lib_1.12.1
[135] Cairo_1.5-12.2 mixtools_1.2.0
[137] RCurl_1.98-1.5 broom_0.7.9
[139] hms_1.1.1 rhdf5_2.34.0
[141] colorspace_2.0-2 BiocManager_1.30.16
[143] ggbeeswarm_0.6.0 shape_1.4.6
[145] sass_0.4.0 Rcpp_1.0.7
[147] mclust_5.4.7 RANN_2.6.1
[149] mvtnorm_1.1-3 fansi_0.5.0
[151] parallelly_1.28.1 ggridges_0.5.3
[153] lifecycle_1.0.1 bluster_1.0.0
[155] minqa_1.2.4 testthat_3.1.0
[157] leiden_0.3.8 jquerylib_0.1.4
[159] snakecase_0.11.0 desc_1.4.0
[161] RcppAnnoy_0.0.19 TMB_1.7.22
[163] htmlwidgets_1.5.4 beachmat_2.6.4
[165] polyclip_1.10-0 purrr_0.3.4
[167] mgcv_1.8-38 globals_0.14.0
[169] leidenAlg_0.1.1 codetools_0.2-18
[171] lubridate_1.8.0 gtools_3.9.2
[173] prettyunits_1.1.1 R.methodsS3_1.8.1
[175] gtable_0.3.0 DBI_1.1.1
[177] git2r_0.28.0 tensor_1.5
[179] httr_1.4.2 highr_0.9
[181] KernSmooth_2.23-20 stringi_1.7.4
[183] progress_1.2.2 farver_2.1.0
[185] reshape2_1.4.4 annotate_1.68.0
[187] hexbin_1.28.2 colorRamps_2.3
[189] sccore_1.0.0 boot_1.3-28
[191] grr_0.9.5 BiocNeighbors_1.8.2
[193] lme4_1.1-27.1 geneplotter_1.68.0
[195] scattermore_0.7 DESeq2_1.30.1
[197] bit_4.0.4 spatstat.data_2.1-0
[199] janitor_2.1.0 pkgconfig_2.0.3
[201] lmerTest_3.1-3 knitr_1.36