Batch correction with conos
Will Macnair
Institute for Molecular Life Sciences, University of Zurich, SwitzerlandSwiss Institute of Bioinformatics (SIB), University of Zurich, SwitzerlandApril 29, 2021
Last updated: 2021-04-29
Checks: 6 1
Knit directory: MS_lesions/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
The global environment had objects present when the code in the R Markdown file was run. These objects can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment. Use wflow_publish or wflow_build to ensure that the code is always run in an empty environment.
The following objects were defined in the global environment when these results were created:
| Name | Class | Size |
|---|---|---|
| q | function | 1008 bytes |
The command set.seed(20210118) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version a6bdc98. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rprofile
Ignored: .Rproj.user/
Ignored: ._.DS_Store
Ignored: ._MS_lesions.sublime-project
Ignored: MS_lesions.sublime-project
Ignored: MS_lesions.sublime-workspace
Ignored: analysis/.__site.yml
Ignored: analysis/ms02_doublet_id_cache/
Ignored: analysis/ms03_SampleQC_cache/
Ignored: analysis/ms05_splitting_cache/
Ignored: analysis/ms07_soup_cache/
Ignored: analysis/ms08_modules_cache/
Ignored: analysis/ms10_muscat_run01_cache/
Ignored: analysis/ms10_muscat_run02_cache/
Ignored: analysis/ms10_muscat_template_cache/
Ignored: analysis/supp10_muscat_cache/
Ignored: data/
Ignored: output/
Untracked files:
Untracked: analysis/ms10_muscat_run01.Rmd
Untracked: analysis/ms10_muscat_run02.Rmd
Untracked: analysis/ms10_muscat_template.Rmd
Untracked: code/ms10_muscat_fns.R
Untracked: code/ms10_muscat_runs.R
Untracked: code/muscat_plan.txt
Untracked: code/plot_dotplot.R
Untracked: code/supp10_muscat.R
Unstaged changes:
Modified: analysis/index.Rmd
Modified: analysis/ms03_SampleQC.Rmd
Modified: analysis/ms07_soup.Rmd
Modified: analysis/ms08_modules.Rmd
Modified: analysis/supp10_muscat.Rmd
Modified: code/ms00_utils.R
Modified: code/ms03_SampleQC.R
Modified: code/ms07_soup.R
Deleted: code/ms10_muscat.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ms04_conos.Rmd) and HTML (docs/ms04_conos.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | a6221cc | wmacnair | 2021-04-07 | Giant update of recent work |
| Rmd | b7ecf3b | wmacnair | 2021-02-15 | Finalized cluster splitting and merging, did modules |
| Rmd | fd8a5bc | wmacnair | 2021-02-10 | Running and checking conos outputs |
Setup / definitions
Libraries
Helper functions
source('code/ms00_utils.R')
source('code/ms03_SampleQC.R')
source('code/ms04_conos.R')Inputs
sce_f = 'data/sce_raw/ms_sce.rds'
meta_f = 'data/metadata/metadata_updated_20201127.txt'
# qc info
dbl_dir = 'output/ms02_doublet_id'
dbl_f = file.path(dbl_dir, 'scDblFinder_outputs.txt')
loom_dir = 'data/loom_files'
qc_dir = 'output/ms03_SampleQC'
qc_f = file.path(qc_dir, 'ms_qc_dt.txt')
keep_f = sprintf('%s/keep_dt_%s.txt', qc_dir, '2021-02-10')Outputs
# define save directory
save_dir = 'output/ms04_conos'
date_tag = '2021-02-11'
if (!dir.exists(save_dir))
dir.create(save_dir)
# define conos run
res_list = c(1, 2, 4, 6, 8)
sel_res = 8
min_cells = 100
conos_f = sprintf('%s/conos_clusts_%s.txt', save_dir, date_tag)
viz_f = sprintf('%s/conos_viz_%s.txt', save_dir, date_tag)
# umap_f = sprintf('%s/conos_umap_%s.txt', save_dir, date_tag)
n_per_conos = 200
graph_f = sprintf('%s/conos_graph_%s.txt', save_dir, date_tag)
umap_many_f = sprintf('%s/conos_umap_sub_%s.txt', save_dir, date_tag)
spread_list = c(1, 2, 4, 8)
min_list = c(1e-2, 1e-1, 1e-0)
# define file to save these
prior_f = sprintf('%s/prior_marker_means_%s.txt', save_dir, date_tag)
# define outputs
annot_f = sprintf('%s/conos_annot_%s.txt', save_dir, date_tag)
# define list of groups to use
fm_groups = list(
oligo = "Oligodendrocytes",
opc = "Oligodendrocyte precursor cells",
astro = "Astrocytes",
micro_immune = c("Microglia", "Plasma B cells"),
excit_neuron = "Excitatory neurons",
inhib_neuron = "Inhibitory neurons",
endo_peri = c("Endothelial cells", "Pericytes")
)
fm_pat = sprintf("%s/fm_%s_%s_%s.txt", save_dir, '%s', '%s', date_tag)
n_cells = 500Load inputs
dbl_dt = dbl_f %>% fread %>%
.[, .(cell_id, cxds = pmax(log10(cxds_score), -5), dbl_logscore = log10(score))]
qc_dt = get_qc_dt(qc_dir, qc_f) %>%
merge(dbl_dt, by = 'cell_id')meta_dt = meta_f %>% fread %>%
.[, .(sample_id = library_id, patient_id)]keep_dt = keep_f %>% fread
calc_conos(sce_f, save_dir, date_tag, keep_dt$cell_id, res_list,
n_cores = 48)done!NULLconos_dt = load_conos_dt(conos_f, sel_res, min_cells)umap_dt = viz_f %>% fread %>%
set_colnames(c('cell_id', 'umap1', 'umap2'))prior_dt = get_prior_dt()Processing / calculations
# extract counts, means
marker_list = prior_dt$symbol
marker_exp_dt = calc_marker_exp_dt(marker_list, sce_f, prior_f, qc_f, conos_dt,
cell_ids = conos_dt$cell_id) %>%
merge(prior_dt, by = 'symbol')already done! loadinglabels_dt = calc_labels_dt(marker_exp_dt)
marker_exp_dt = merge(marker_exp_dt, labels_dt, by = 'conos')
conos_dt = merge(conos_dt, labels_dt, by = 'conos')tests = c('binom', 'wilcox', 't')
for (n in names(fm_groups)) {
# define filename
fm_fs = tests %>% sapply(function(t)
sprintf(fm_pat, n, t) %>% file.path(save_dir, .))
broad_types = fm_groups[[n]]
# run findMarkers
calc_find_markers_broad(sce_f, fm_fs, n, broad_types, tests,
conos_dt, n_cells, n_cores = 8)
}set.seed(20210212)
conos_sub = calc_conos_sub(conos_dt, n_per_conos)
umap_all = calc_umap_sub(conos_sub, graph_f, umap_many_f, spread_list, min_list,
seed = 20210212)set.seed(20210212)
umap_sub = umap_dt[ sample(nrow(umap_dt), 2e4) ] %>%
merge(conos_dt, by='cell_id')
umap_qc_dt = umap_sub[, .(cell_id, umap1, umap2)] %>%
merge(qc_dt, by='cell_id') %>%
melt(id=c('cell_id', 'sample_id', 'umap1', 'umap2'),
variable.name = 'qc_var', value.name = 'qc_val')Analysis
Density over UMAP
# plot densities
(plot_umap_dens(umap_dt))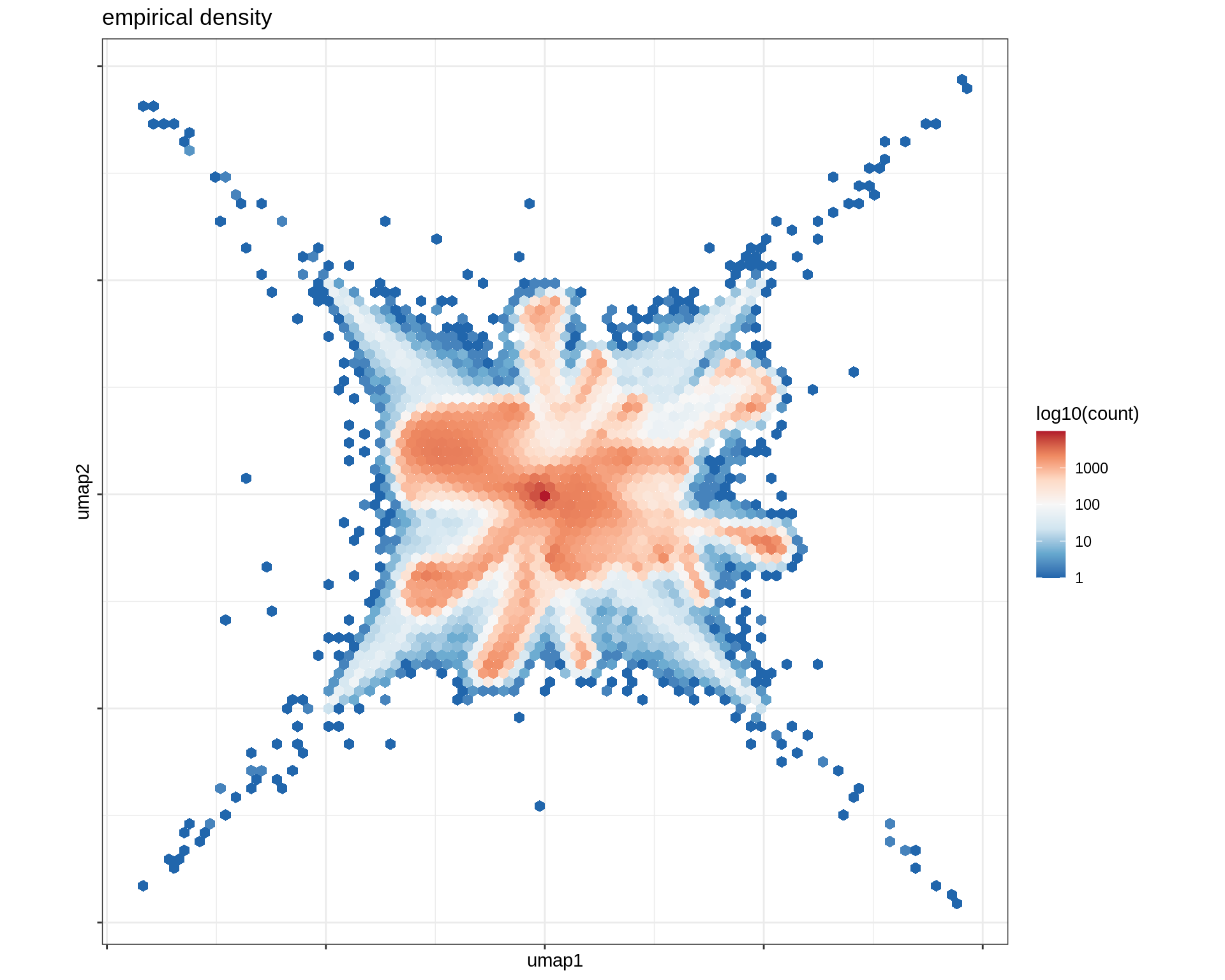
Broad celltypes over UMAP
(plot_umap_broad(umap_sub))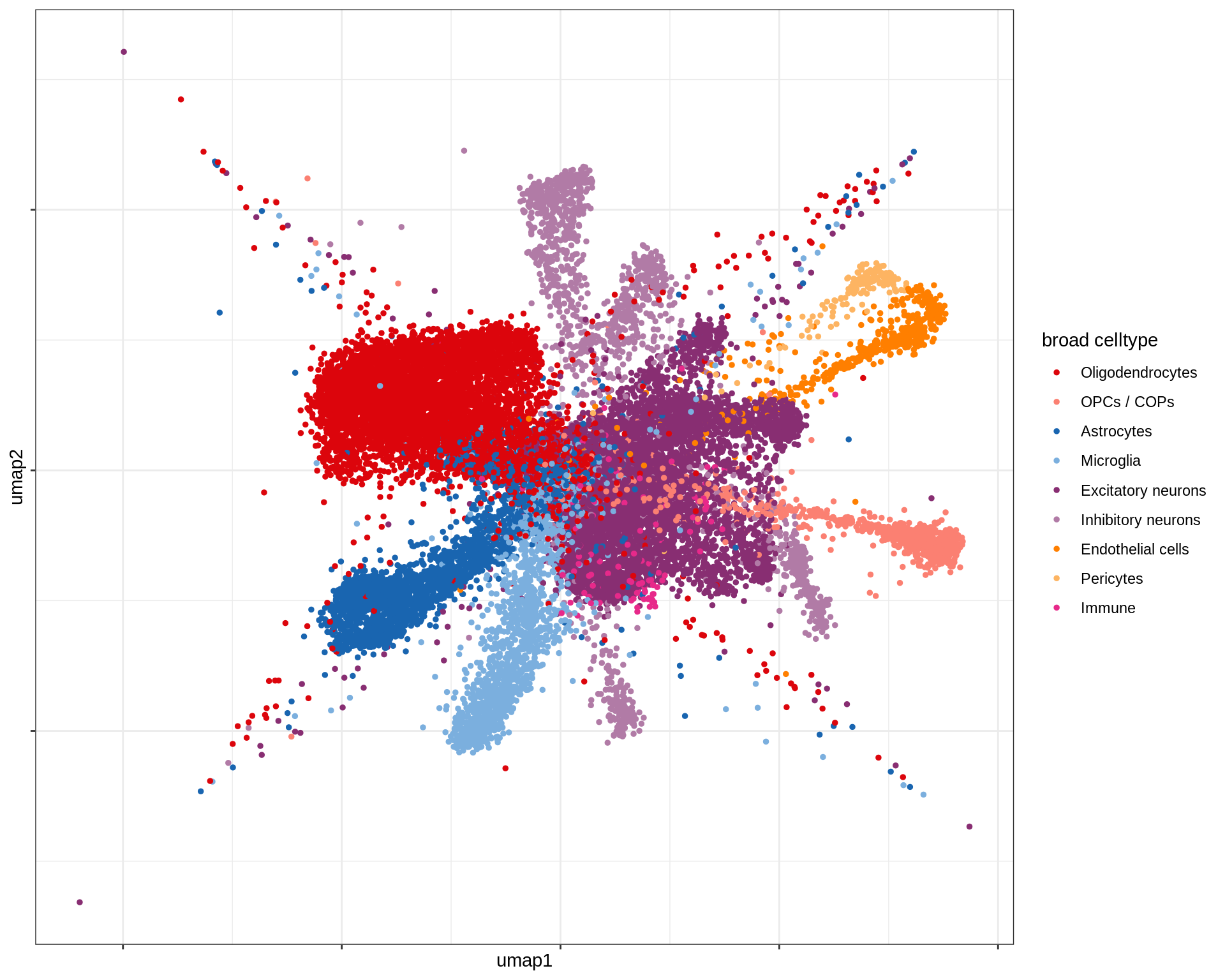
Fine celltypes over UMAP
set.seed(20210202)
for (b in broad_ord) {
cat('### ', b, ' \n')
print(plot_umap_split(conos_dt, umap_dt, b))
cat('\n\n')
}Oligodendrocytes
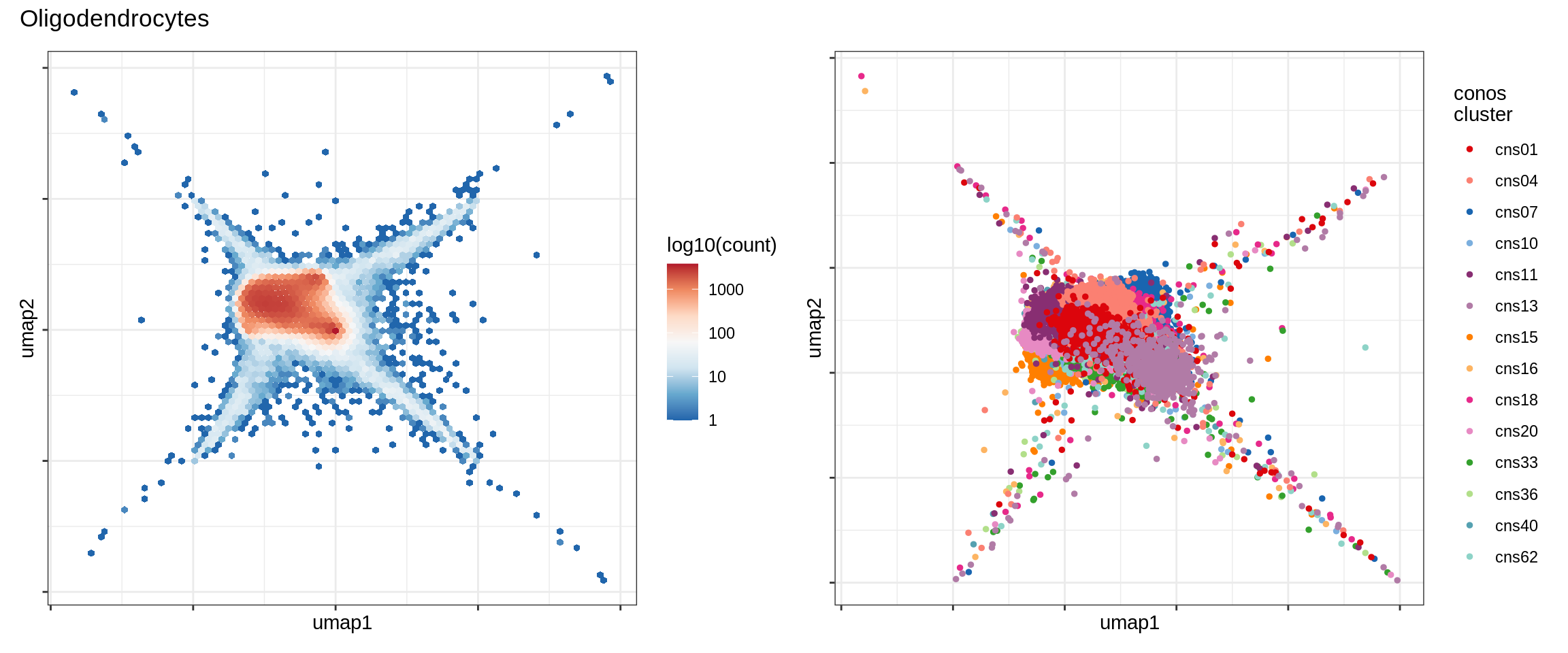
OPCs / COPs
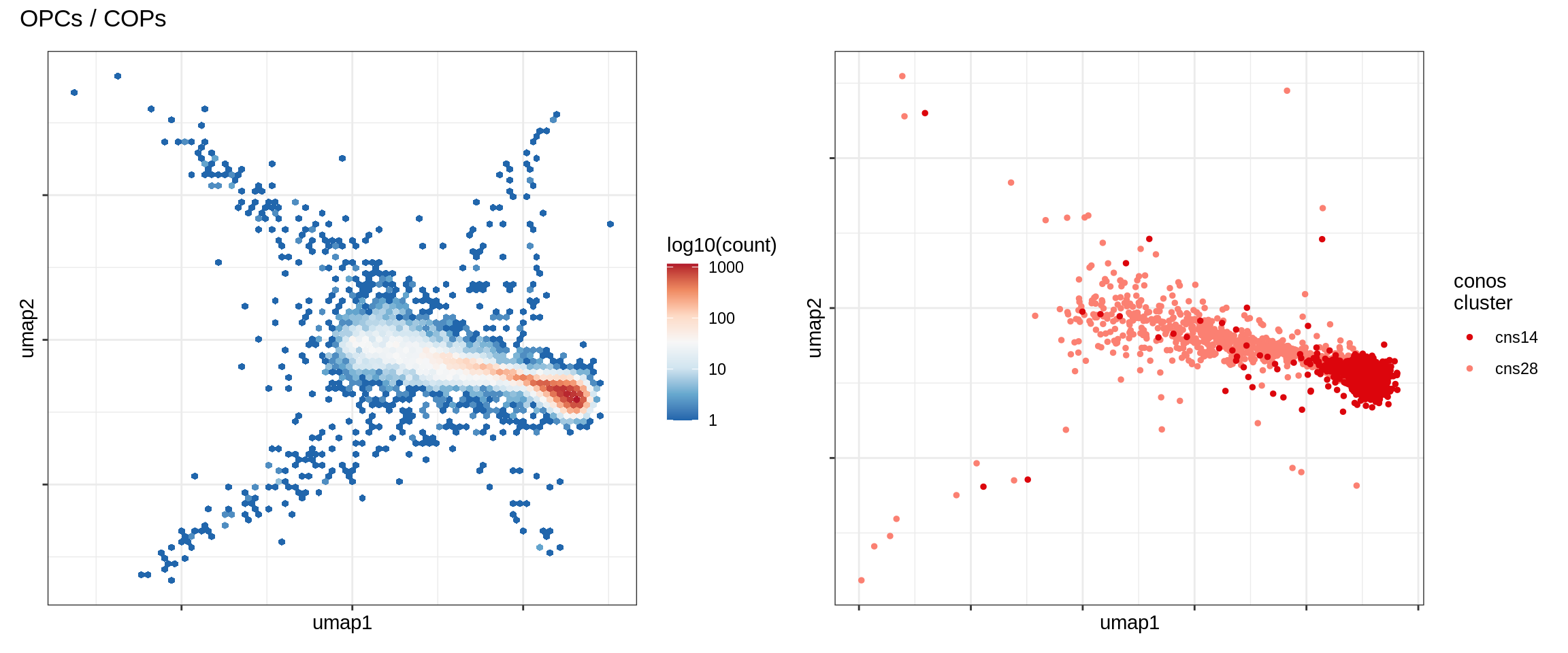
Astrocytes
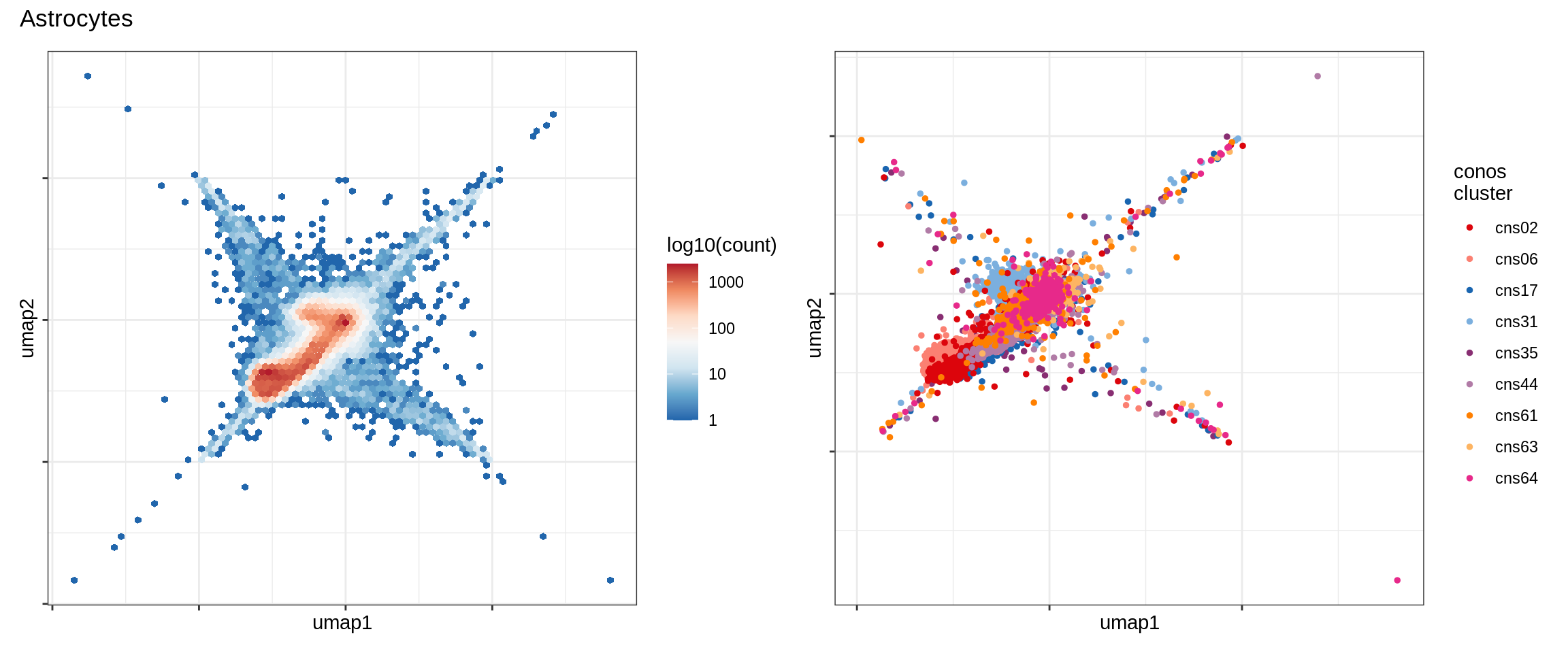
Microglia
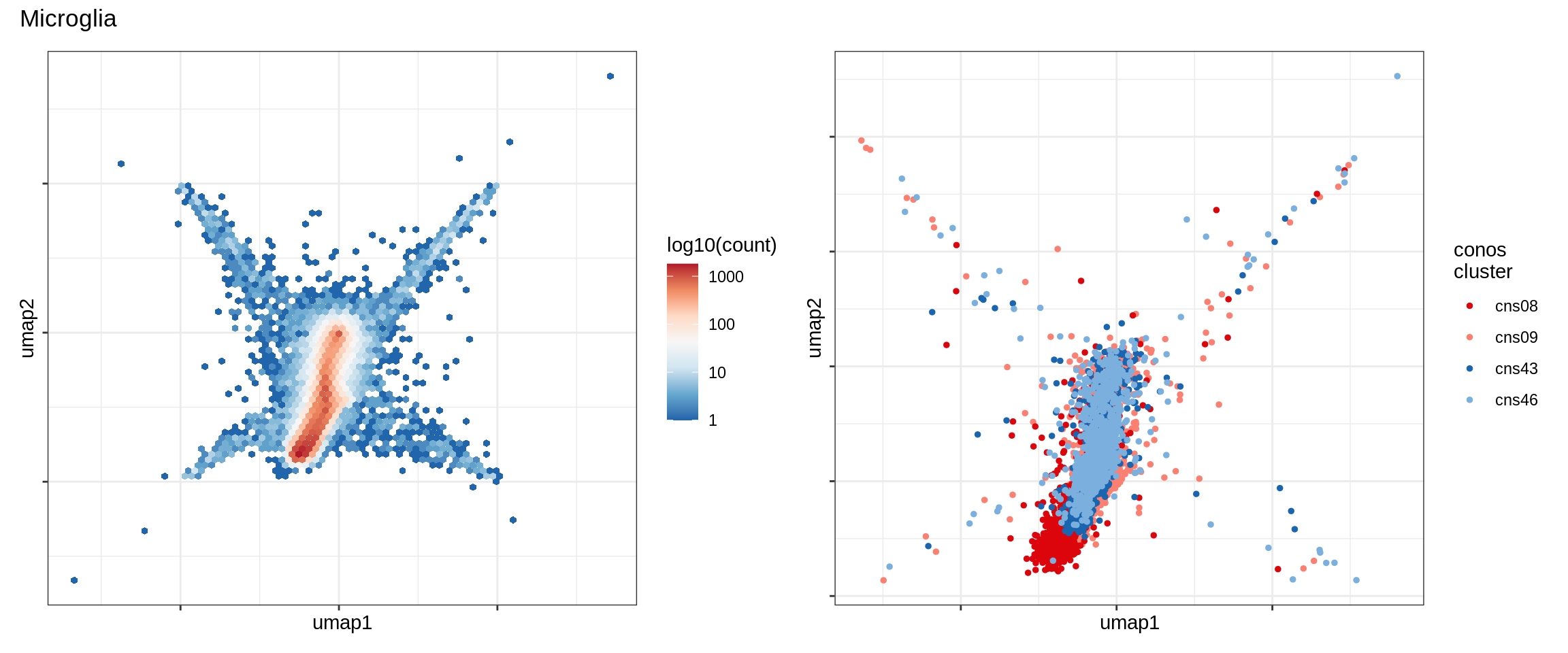
Excitatory neurons
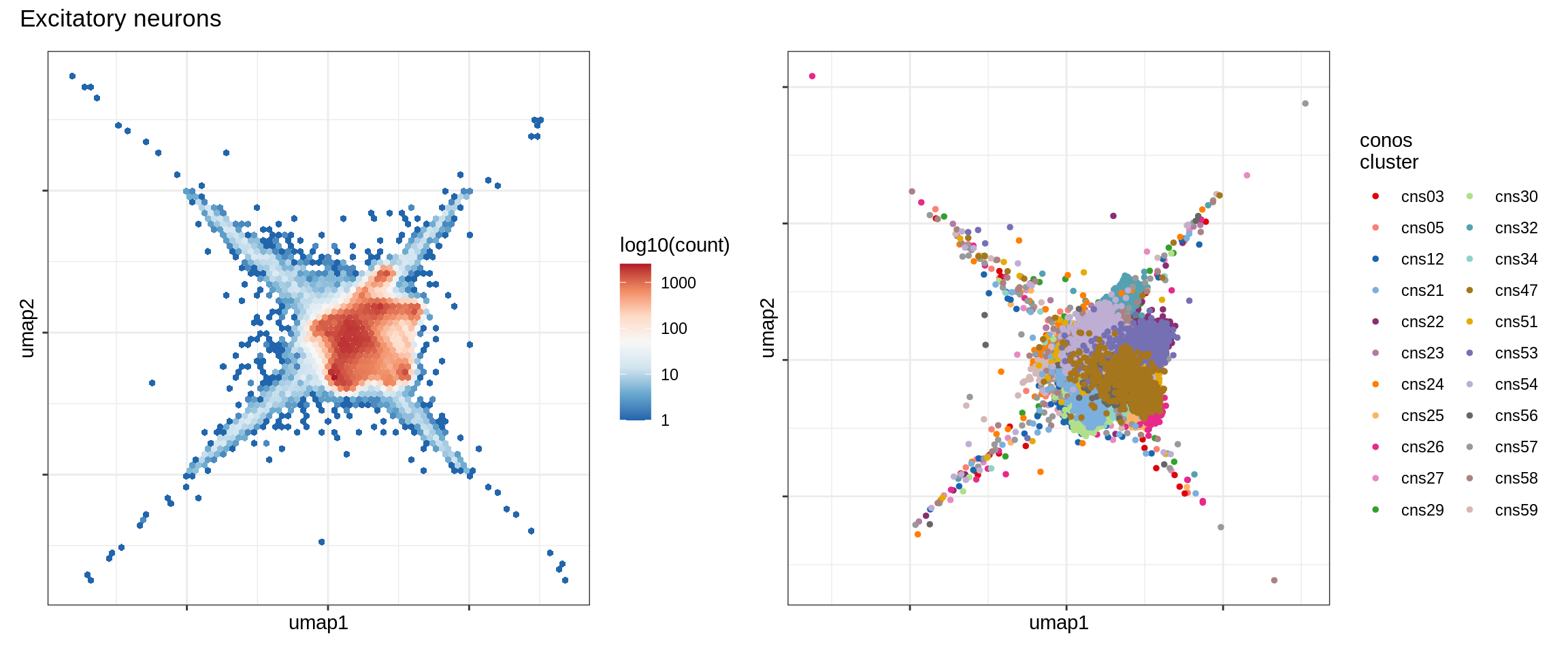
Inhibitory neurons
Endothelial cells
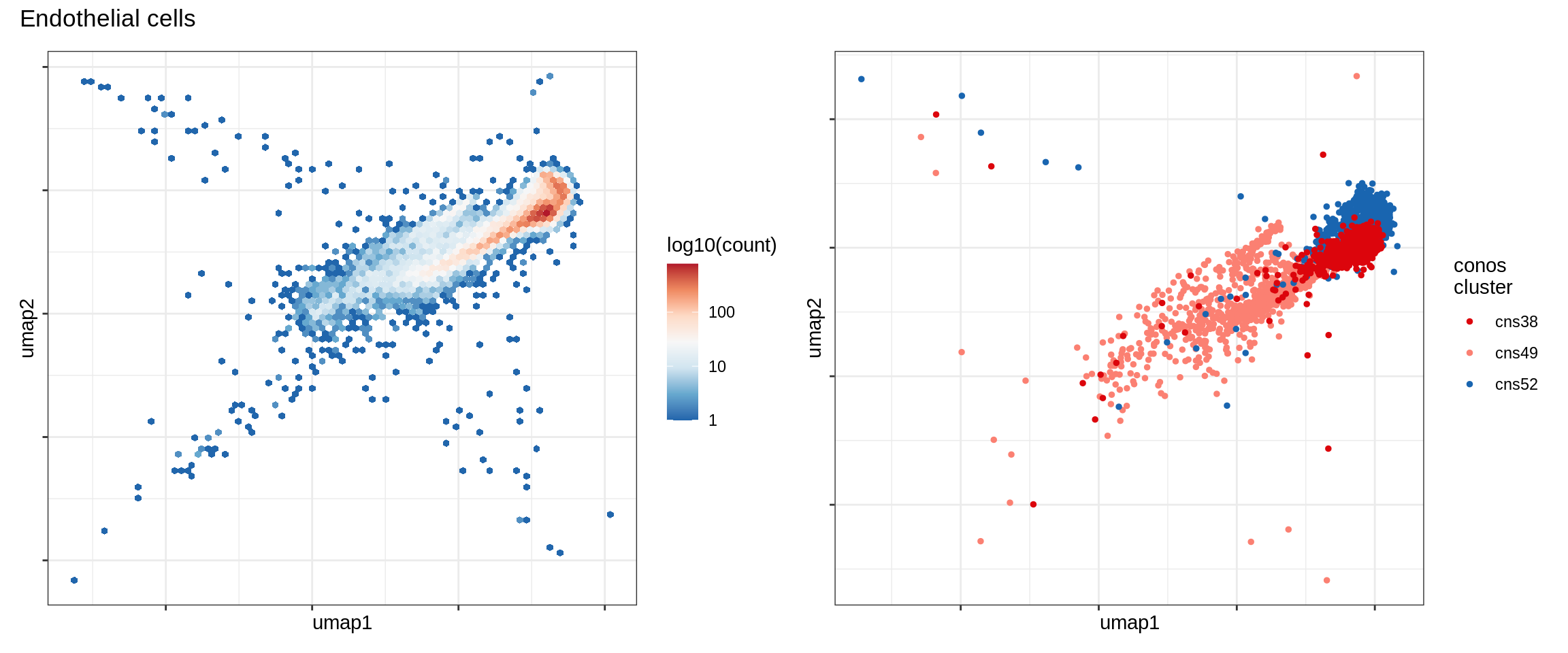
Pericytes
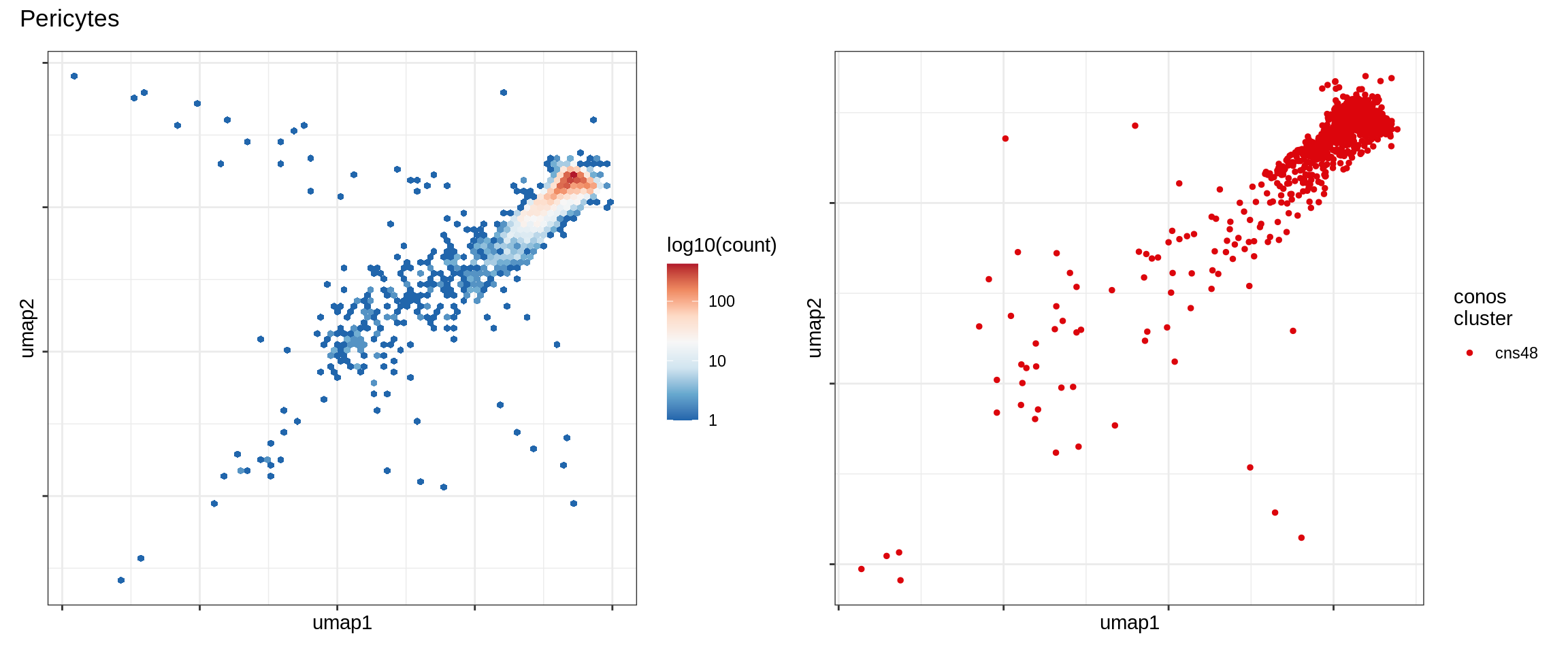
Immune
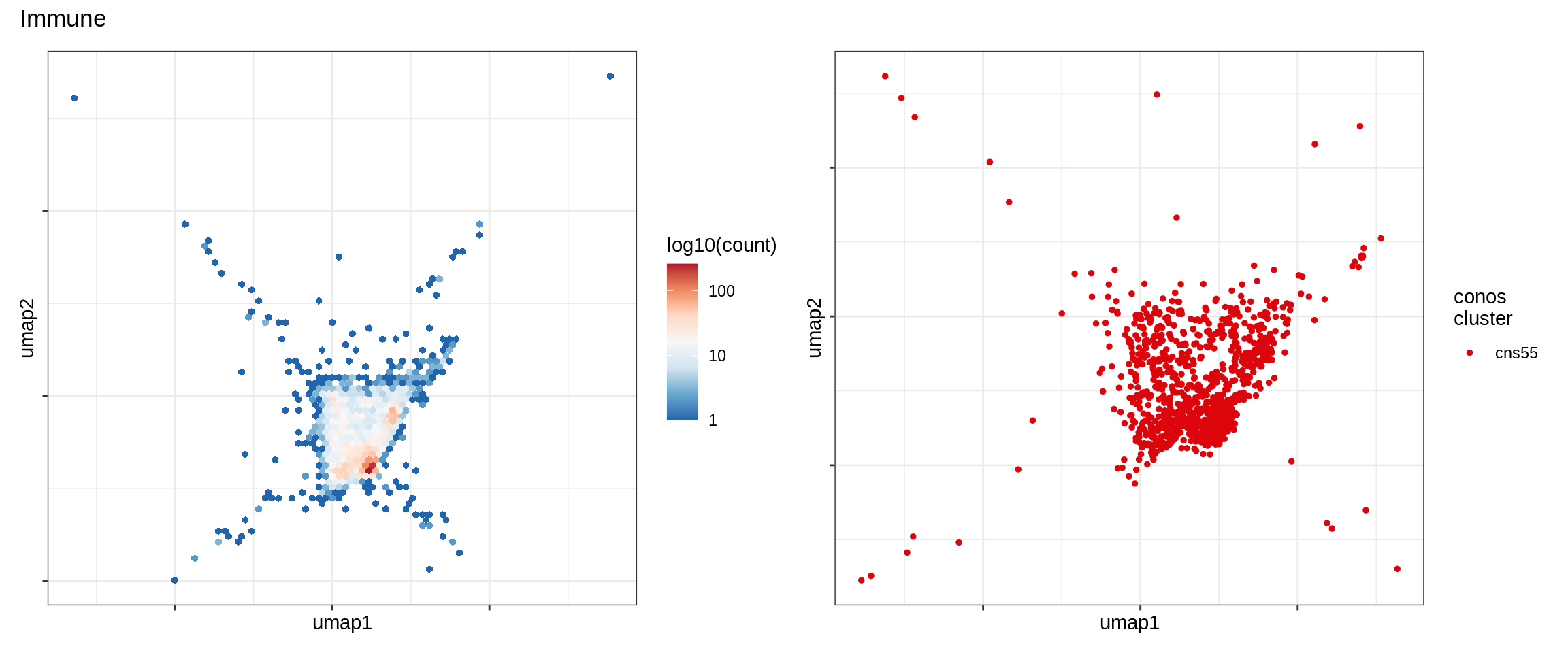
Expression of known marker genes
draw(marker_heatmap_fn(marker_exp_dt), merge_legend = TRUE,
heatmap_legend_side = "bottom", annotation_legend_side = "bottom")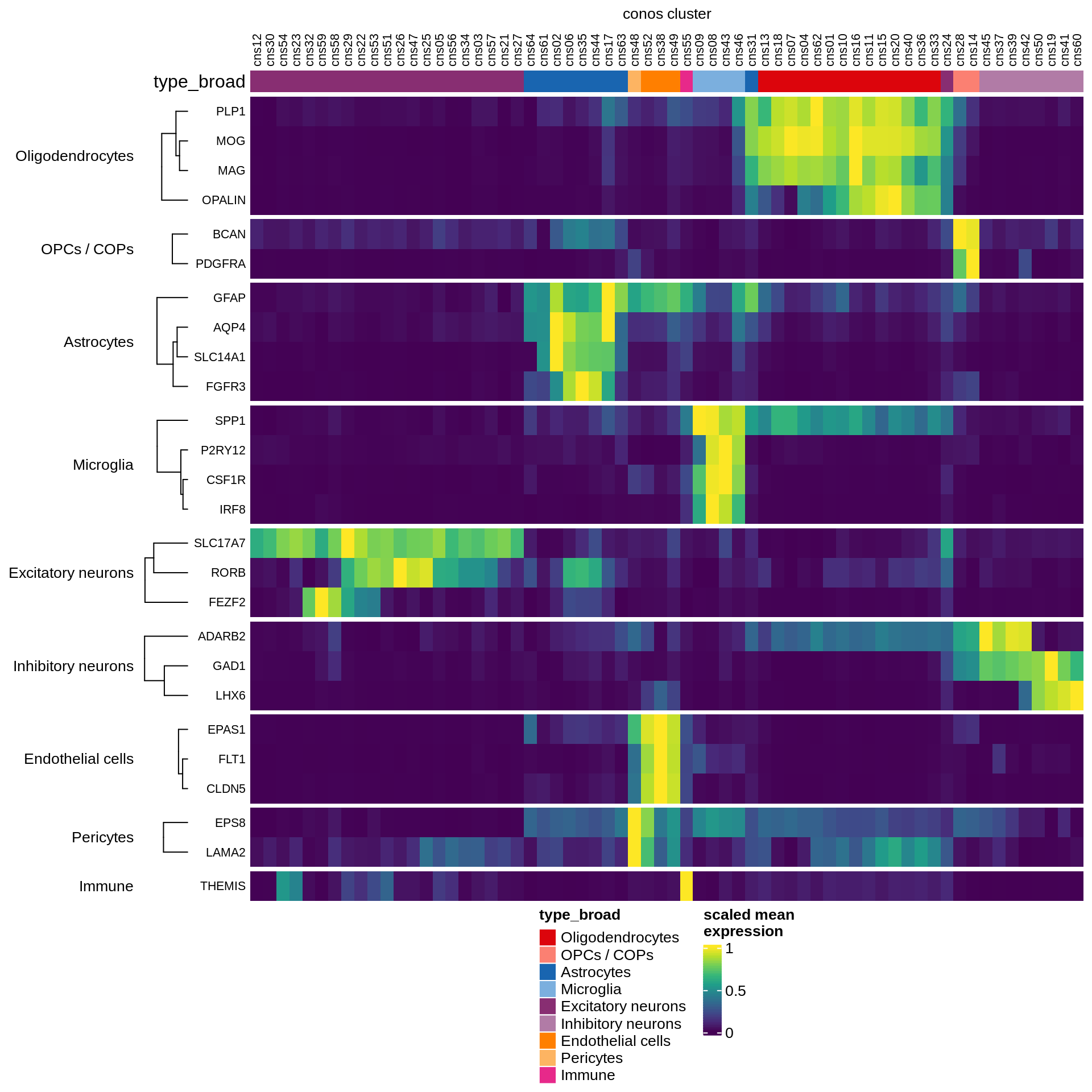
Celltype mixing across samples
(plot_conos_mixing(conos_dt, meta_dt))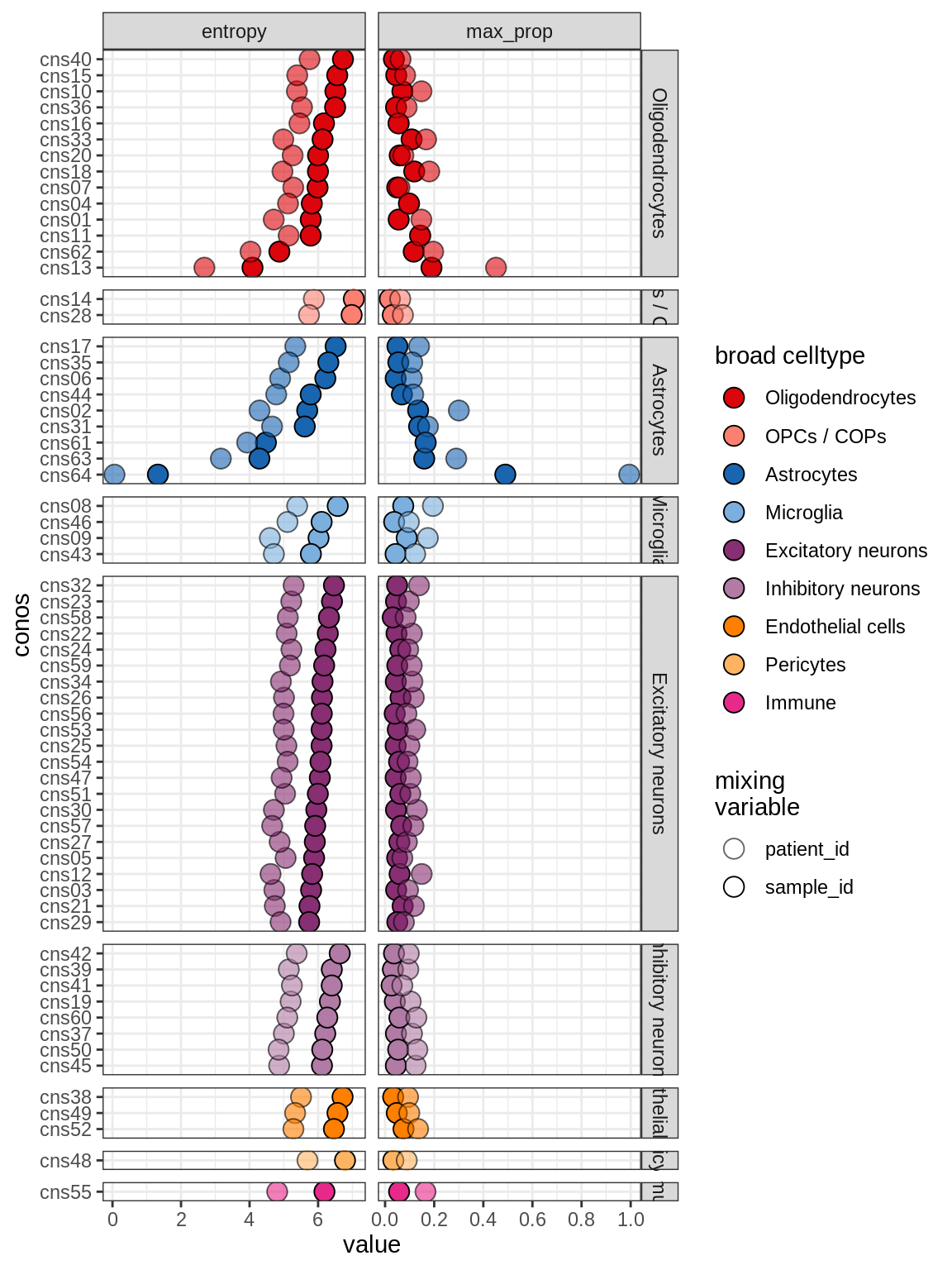
QC metrics split by conos celltype
(plot_qc_violins(qc_dt, conos_dt))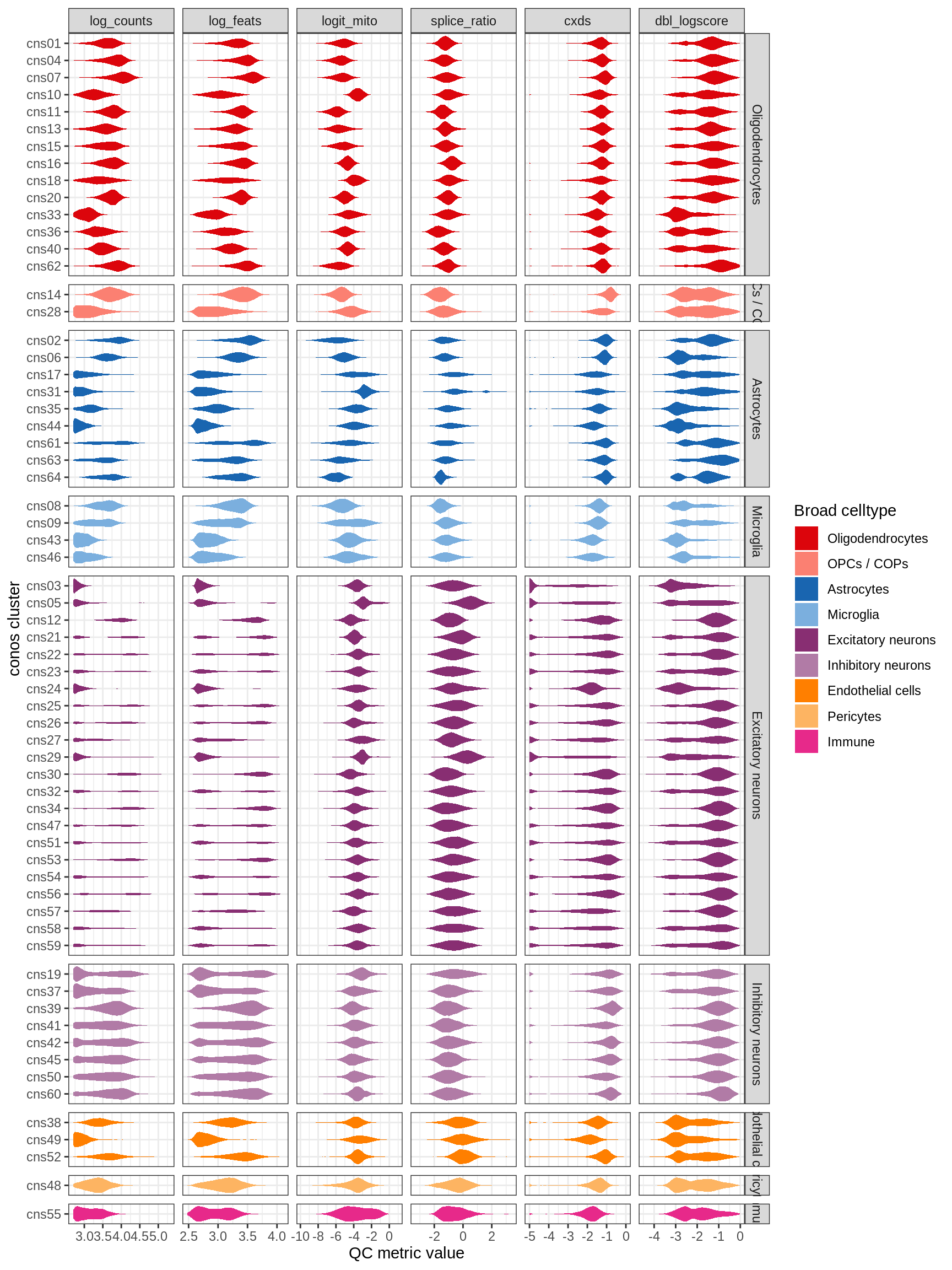
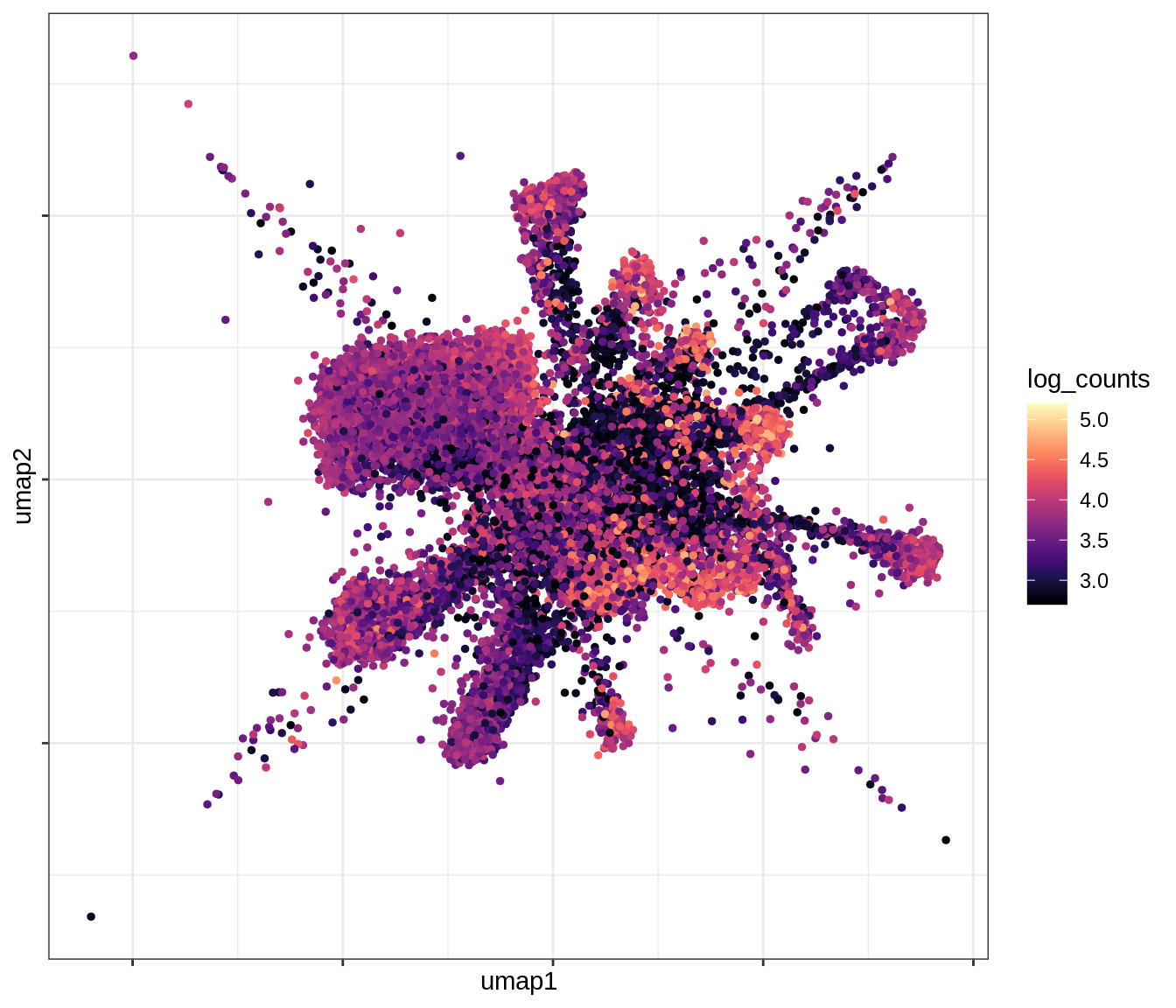
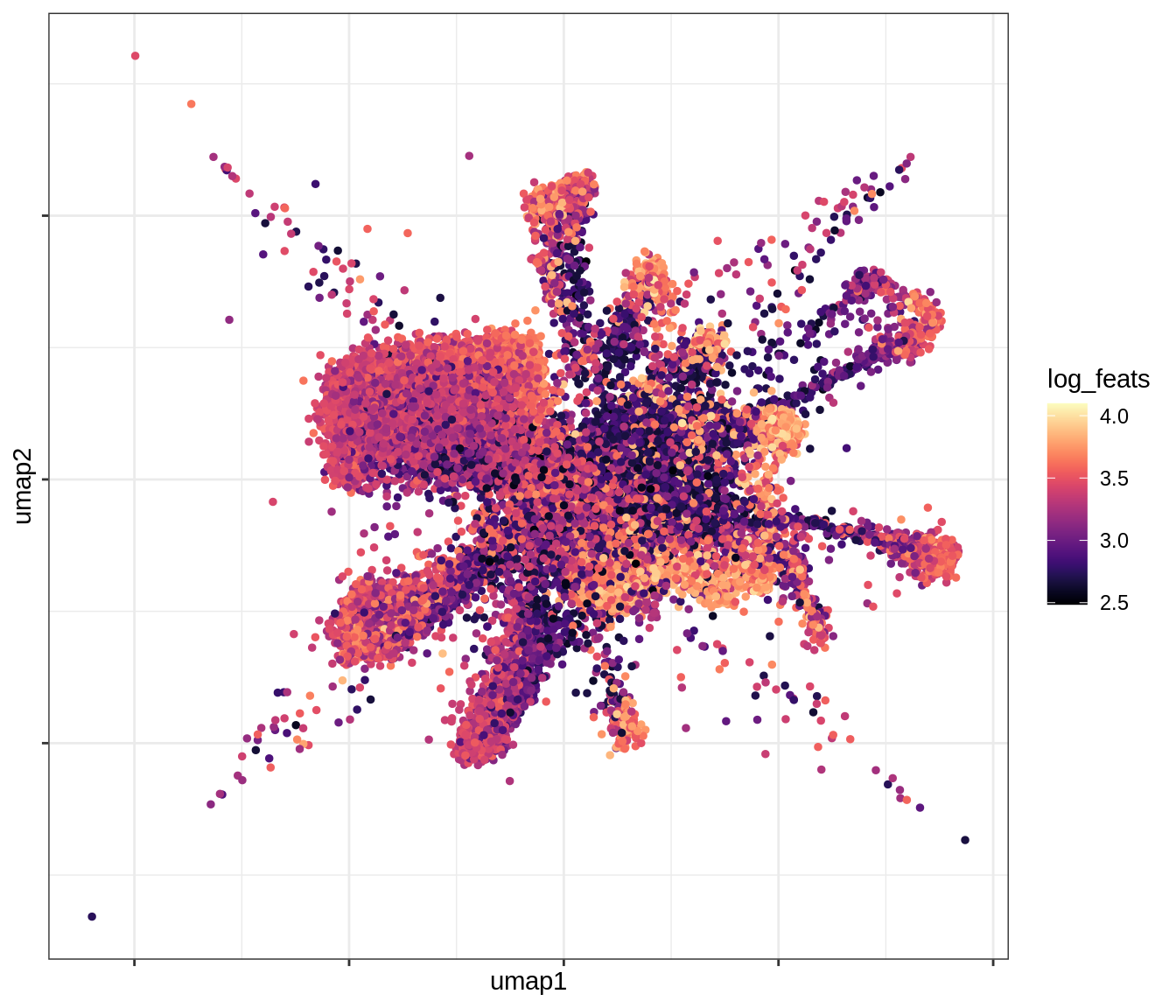
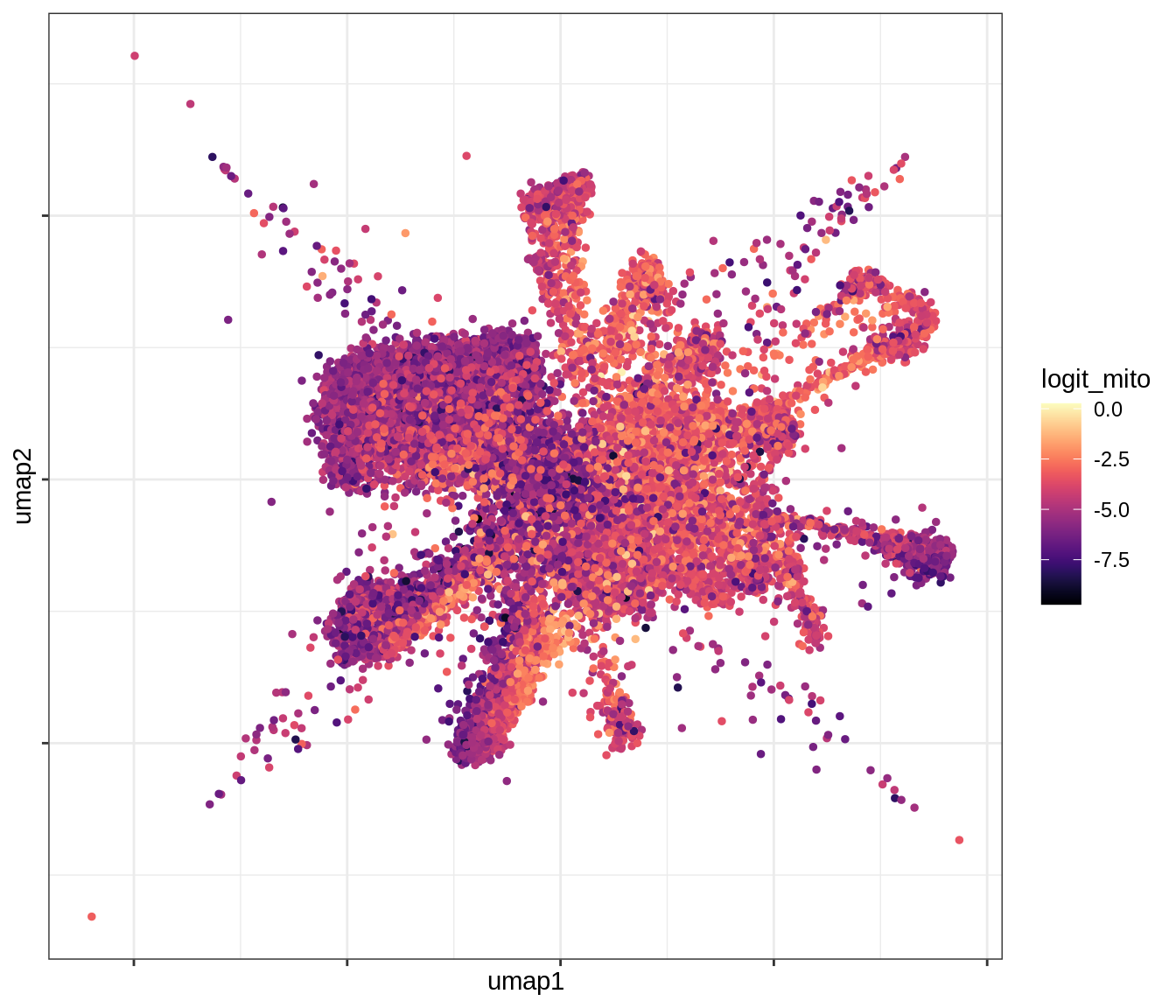
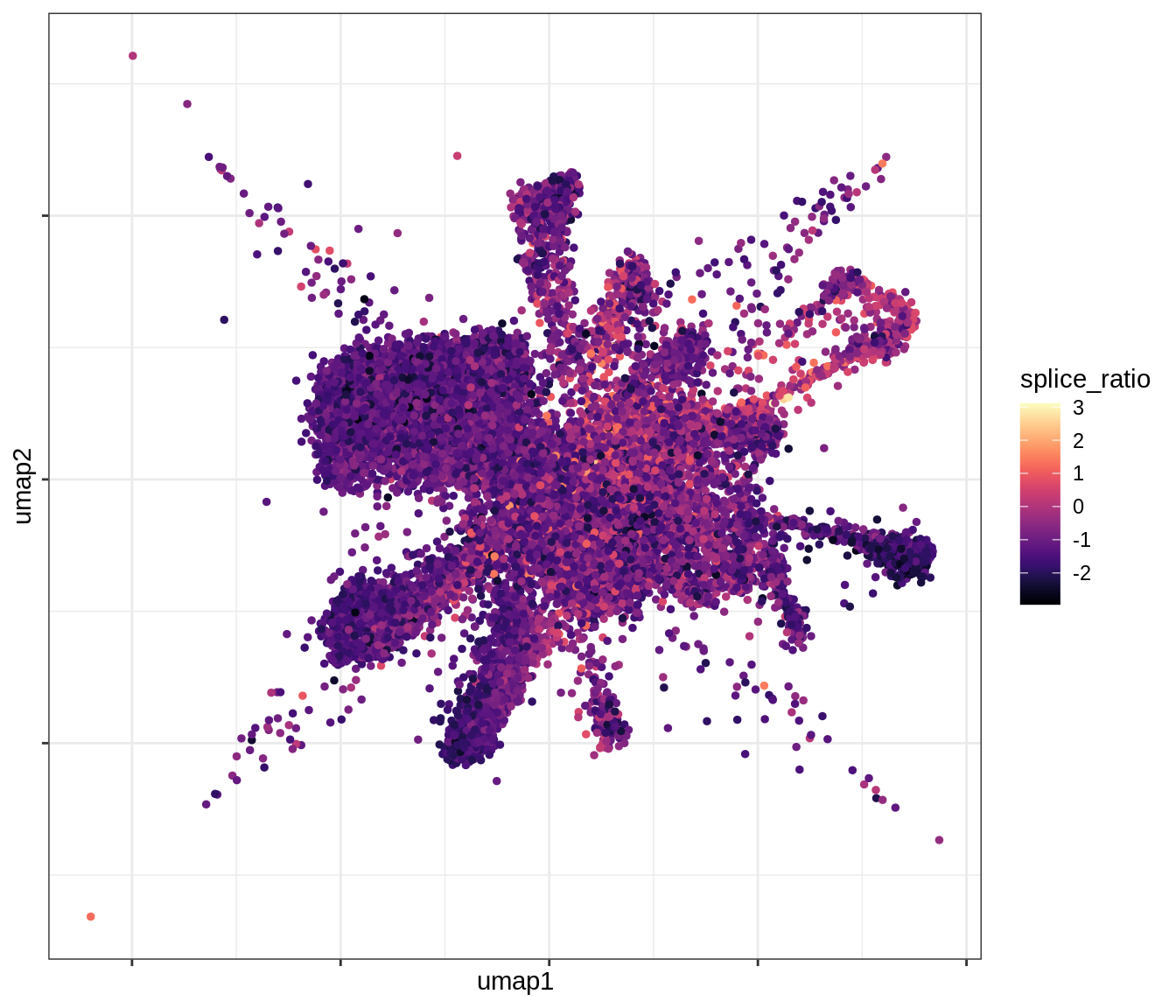
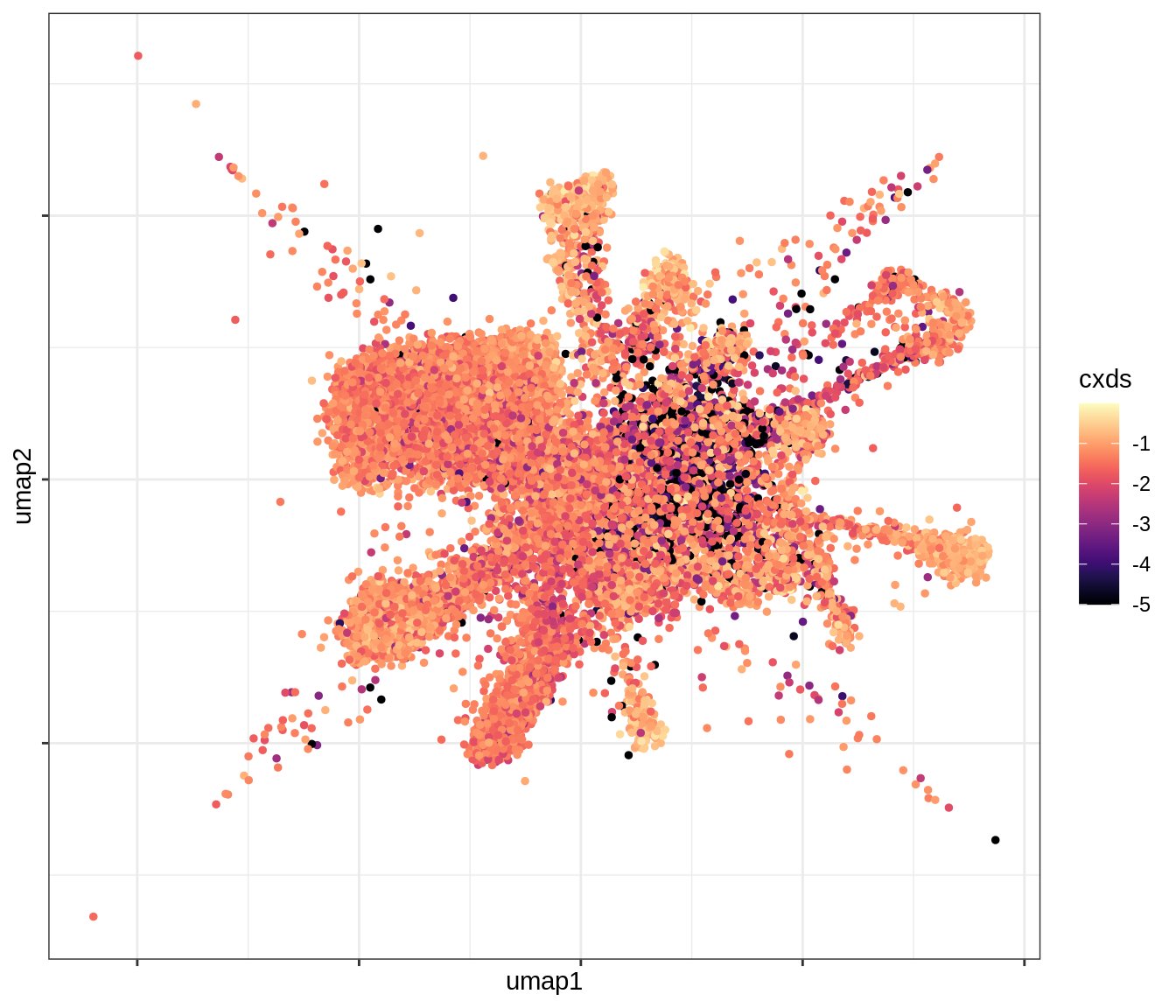
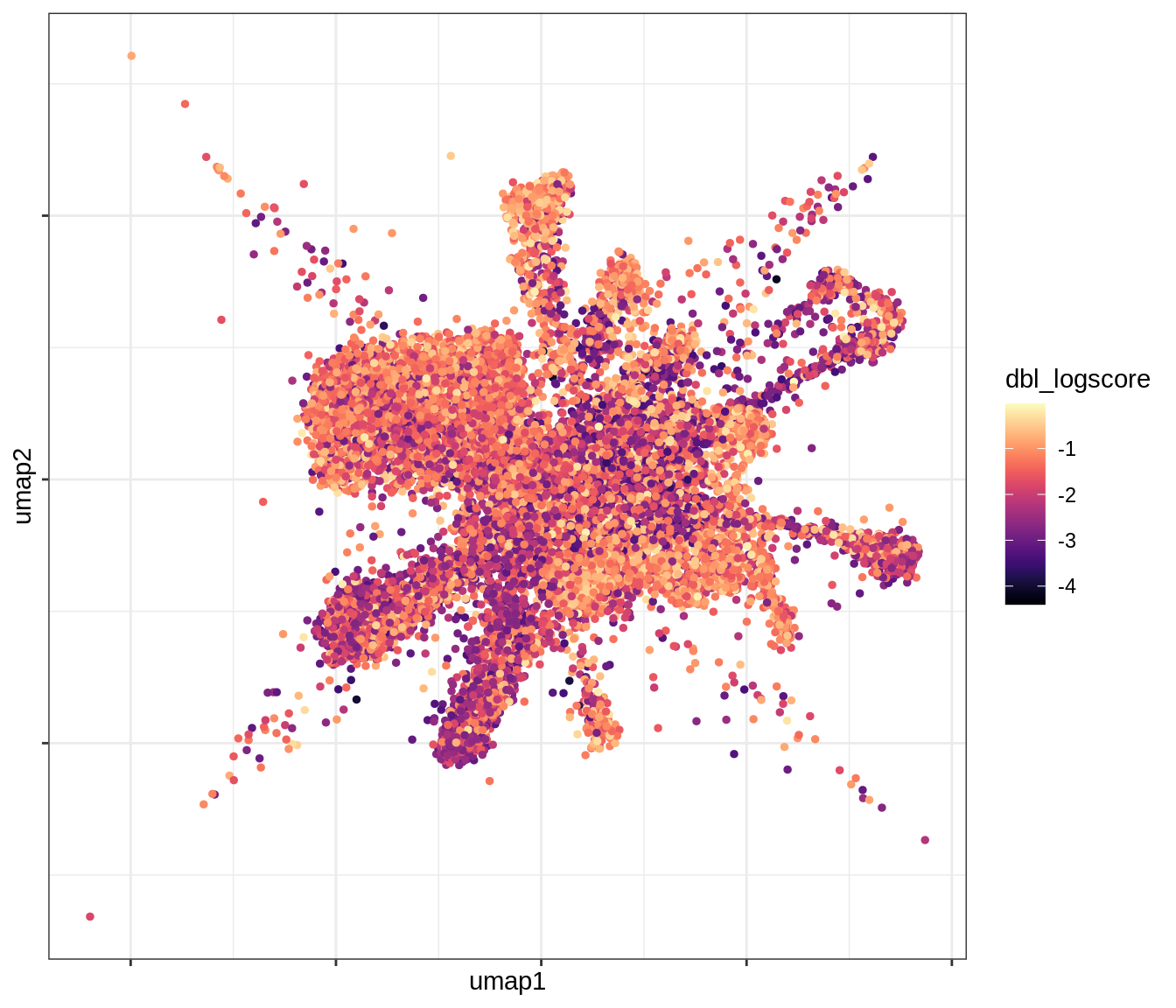
QC metrics celltypes over UMAP
for (v in levels(umap_qc_dt$qc_var)) {
cat('### ', v, '\n')
print(plot_umap_qc(umap_qc_dt, v))
cat('\n\n')
}log_counts
log_feats
logit_mito
splice_ratio
cxds
dbl_logscore
Outputs
conos_dt %>% fwrite(file = annot_f)devtools::session_info()Registered S3 method overwritten by 'cli':
method from
print.boxx spatstat.geom- Session info ---------------------------------------------------------------
setting value
version R version 4.0.3 (2020-10-10)
os CentOS Linux 7 (Core)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_US.UTF-8
ctype C
tz Europe/Zurich
date 2021-04-29
- Packages -------------------------------------------------------------------
package * version date lib
abind 1.4-5 2016-07-21 [2]
assertthat * 0.2.1 2019-03-21 [2]
beachmat 2.6.4 2020-12-20 [1]
beeswarm 0.3.1 2021-03-07 [1]
Biobase * 2.50.0 2020-10-27 [1]
BiocGenerics * 0.36.1 2021-04-16 [1]
BiocManager 1.30.12 2021-03-28 [1]
BiocNeighbors 1.8.2 2020-12-07 [1]
BiocParallel * 1.24.1 2020-11-06 [1]
BiocSingular 1.6.0 2020-10-27 [1]
BiocStyle * 2.18.1 2020-11-24 [1]
bit 4.0.4 2020-08-04 [2]
bit64 4.0.5 2020-08-30 [2]
bitops 1.0-6 2013-08-17 [2]
bluster 1.0.0 2020-10-27 [1]
bslib 0.2.4 2021-01-25 [2]
cachem 1.0.4 2021-02-13 [2]
Cairo 1.5-12.2 2020-07-07 [2]
callr 3.6.0 2021-03-28 [2]
circlize * 0.4.12 2021-01-08 [1]
cli 2.4.0 2021-04-05 [2]
clue 0.3-59 2021-04-16 [1]
cluster 2.1.2 2021-04-17 [2]
codetools 0.2-18 2020-11-04 [2]
colorout * 1.2-2 2021-04-15 [1]
colorspace 2.0-0 2020-11-11 [2]
ComplexHeatmap * 2.6.2 2020-11-12 [1]
conos * 1.4.0 2021-02-23 [1]
cowplot 1.1.1 2020-12-30 [2]
crayon 1.4.1 2021-02-08 [2]
data.table * 1.14.0 2021-02-21 [2]
DBI 1.1.1 2021-01-15 [2]
DelayedArray 0.16.3 2021-03-24 [1]
DelayedMatrixStats 1.12.3 2021-02-03 [1]
deldir 0.2-10 2021-02-16 [2]
desc 1.3.0 2021-03-05 [2]
devtools 2.4.0 2021-04-07 [1]
digest 0.6.27 2020-10-24 [2]
dplyr 1.0.5 2021-03-05 [2]
dqrng 0.2.1 2019-05-17 [2]
edgeR 3.32.1 2021-01-14 [1]
ellipsis 0.3.1 2020-05-15 [2]
evaluate 0.14 2019-05-28 [2]
fansi 0.4.2 2021-01-15 [2]
farver 2.1.0 2021-02-28 [2]
fastmap 1.1.0 2021-01-25 [2]
fitdistrplus 1.1-3 2020-12-05 [2]
forcats * 0.5.1 2021-01-27 [2]
foreach 1.5.1 2020-10-15 [2]
fs 1.5.0 2020-07-31 [2]
future 1.21.0 2020-12-10 [2]
future.apply 1.7.0 2021-01-04 [2]
generics 0.1.0 2020-10-31 [2]
GenomeInfoDb * 1.26.7 2021-04-08 [1]
GenomeInfoDbData 1.2.4 2021-04-15 [1]
GenomicRanges * 1.42.0 2020-10-27 [1]
GetoptLong 1.0.5 2020-12-15 [1]
ggbeeswarm 0.6.0 2017-08-07 [1]
ggplot.multistats * 1.0.0 2019-10-28 [1]
ggplot2 * 3.3.3 2020-12-30 [2]
ggrepel 0.9.1 2021-01-15 [2]
ggridges 0.5.3 2021-01-08 [2]
git2r 0.28.0 2021-01-10 [1]
GlobalOptions 0.1.2 2020-06-10 [1]
globals 0.14.0 2020-11-22 [2]
glue 1.4.2 2020-08-27 [2]
goftest 1.2-2 2019-12-02 [2]
gridExtra 2.3 2017-09-09 [2]
grr 0.9.5 2016-08-26 [1]
gtable 0.3.0 2019-03-25 [2]
gtools 3.8.2 2020-03-31 [2]
hdf5r * 1.3.3 2020-08-18 [2]
hexbin 1.28.2 2021-01-08 [2]
highr 0.9 2021-04-16 [2]
htmltools 0.5.1.1 2021-01-22 [2]
htmlwidgets 1.5.3 2020-12-10 [2]
httpuv 1.5.5 2021-01-13 [2]
httr 1.4.2 2020-07-20 [2]
ica 1.0-2 2018-05-24 [2]
igraph * 1.2.6 2020-10-06 [2]
IRanges * 2.24.1 2020-12-12 [1]
irlba 2.3.3 2019-02-05 [2]
iterators * 1.0.13 2020-10-15 [2]
itertools * 0.1-3 2014-03-12 [1]
jquerylib 0.1.3 2020-12-17 [2]
jsonlite 1.7.2 2020-12-09 [2]
kernlab 0.9-29 2019-11-12 [1]
KernSmooth 2.23-18 2020-10-29 [2]
knitr 1.32 2021-04-14 [1]
labeling 0.4.2 2020-10-20 [2]
later 1.1.0.1 2020-06-05 [2]
lattice 0.20-41 2020-04-02 [2]
lazyeval 0.2.2 2019-03-15 [2]
leiden 0.3.7 2021-01-26 [2]
leidenAlg 0.1.1 2021-03-03 [1]
lifecycle 1.0.0 2021-02-15 [2]
limma 3.46.0 2020-10-27 [1]
listenv 0.8.0 2019-12-05 [2]
lmtest 0.9-38 2020-09-09 [2]
locfit 1.5-9.4 2020-03-25 [1]
loomR * 0.2.0 2021-04-15 [1]
magrittr * 2.0.1 2020-11-17 [1]
MASS 7.3-53.1 2021-02-12 [2]
Matrix * 1.3-2 2021-01-06 [2]
Matrix.utils 0.9.8 2020-02-26 [1]
MatrixGenerics * 1.2.1 2021-01-30 [1]
matrixStats * 0.58.0 2021-01-29 [2]
mclust 5.4.7 2020-11-20 [1]
memoise 2.0.0 2021-01-26 [1]
mgcv 1.8-35 2021-04-18 [2]
mime 0.10 2021-02-13 [2]
miniUI 0.1.1.1 2018-05-18 [2]
mixtools 1.2.0 2020-02-07 [1]
munsell 0.5.0 2018-06-12 [2]
mvnfast 0.2.5.1 2020-10-14 [1]
mvtnorm 1.1-1 2020-06-09 [1]
nlme 3.1-152 2021-02-04 [2]
parallelly 1.24.0 2021-03-14 [2]
patchwork * 1.1.1 2020-12-17 [2]
pbapply 1.4-3 2020-08-18 [2]
pillar 1.6.0 2021-04-13 [2]
pkgbuild 1.2.0 2020-12-15 [1]
pkgconfig 2.0.3 2019-09-22 [2]
pkgload 1.2.1 2021-04-06 [2]
plotly 4.9.3 2021-01-10 [2]
plyr 1.8.6 2020-03-03 [2]
png 0.1-7 2013-12-03 [2]
polyclip 1.10-0 2019-03-14 [2]
prettyunits 1.1.1 2020-01-24 [2]
processx 3.5.1 2021-04-04 [2]
promises 1.2.0.1 2021-02-11 [2]
ps 1.6.0 2021-02-28 [2]
purrr 0.3.4 2020-04-17 [2]
R6 * 2.5.0 2020-10-28 [2]
RANN 2.6.1 2019-01-08 [2]
RColorBrewer * 1.1-2 2014-12-07 [2]
Rcpp 1.0.6 2021-01-15 [2]
RcppAnnoy 0.0.18 2020-12-15 [2]
RCurl 1.98-1.3 2021-03-16 [1]
registry 0.5-1 2019-03-05 [1]
remotes 2.3.0 2021-04-01 [1]
reshape2 1.4.4 2020-04-09 [2]
reticulate 1.18 2020-10-25 [2]
rjson 0.2.20 2018-06-08 [1]
rlang 0.4.10 2020-12-30 [2]
rmarkdown 2.7 2021-02-19 [2]
ROCR 1.0-11 2020-05-02 [2]
rpart 4.1-15 2019-04-12 [2]
rprojroot 2.0.2 2020-11-15 [2]
rsvd 1.0.5 2021-04-16 [1]
Rtsne 0.15 2018-11-10 [2]
S4Vectors * 0.28.1 2020-12-09 [1]
SampleQC * 0.4.5 2021-04-15 [1]
sass 0.3.1 2021-01-24 [2]
scales * 1.1.1 2020-05-11 [2]
scater * 1.18.6 2021-02-26 [1]
scattermore 0.7 2020-11-24 [2]
sccore 0.1.2 2021-02-23 [1]
scran * 1.18.7 2021-04-16 [1]
sctransform 0.3.2 2020-12-16 [2]
scuttle 1.0.4 2020-12-17 [1]
segmented 1.3-3 2021-03-08 [1]
seriation * 1.2-9 2020-10-01 [1]
sessioninfo 1.1.1 2018-11-05 [1]
Seurat * 4.0.1 2021-03-18 [2]
SeuratObject * 4.0.0 2021-01-15 [2]
shape 1.4.5 2020-09-13 [2]
shiny 1.6.0 2021-01-25 [2]
SingleCellExperiment * 1.12.0 2020-10-27 [1]
sparseMatrixStats 1.2.1 2021-02-02 [1]
spatstat.core 2.1-2 2021-04-18 [2]
spatstat.data 2.1-0 2021-03-21 [2]
spatstat.geom 2.1-0 2021-04-15 [2]
spatstat.sparse 2.0-0 2021-03-16 [2]
spatstat.utils 2.1-0 2021-03-15 [2]
statmod 1.4.35 2020-10-19 [1]
stringi 1.5.3 2020-09-09 [2]
stringr * 1.4.0 2019-02-10 [2]
SummarizedExperiment * 1.20.0 2020-10-27 [1]
survival 3.2-10 2021-03-16 [2]
tensor 1.5 2012-05-05 [2]
testthat 3.0.2 2021-02-14 [2]
tibble 3.1.1 2021-04-18 [2]
tidyr 1.1.3 2021-03-03 [2]
tidyselect 1.1.0 2020-05-11 [2]
TSP 1.1-10 2020-04-17 [1]
usethis 2.0.1 2021-02-10 [1]
utf8 1.2.1 2021-03-12 [2]
uwot * 0.1.10 2020-12-15 [2]
vctrs 0.3.7 2021-03-29 [2]
vipor 0.4.5 2017-03-22 [1]
viridis * 0.6.0 2021-04-15 [1]
viridisLite * 0.4.0 2021-04-13 [2]
whisker 0.4 2019-08-28 [1]
withr 2.4.2 2021-04-18 [2]
workflowr * 1.6.2 2020-04-30 [1]
xfun 0.22 2021-03-11 [1]
xtable 1.8-4 2019-04-21 [2]
XVector 0.30.0 2020-10-27 [1]
yaml 2.2.1 2020-02-01 [2]
zlibbioc 1.36.0 2020-10-27 [1]
zoo 1.8-9 2021-03-09 [2]
source
CRAN (R 4.0.0)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.3)
Bioconductor
Bioconductor
CRAN (R 4.0.3)
Bioconductor
Bioconductor
Bioconductor
Bioconductor
CRAN (R 4.0.2)
CRAN (R 4.0.2)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Github (jalvesaq/colorout@79931fd)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
Bioconductor
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.2)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Github (mojaveazure/loomR@df0144b)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
Bioconductor
Github (wmacnair/SampleQC@5e3f021)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.2)
CRAN (R 4.0.3)
Bioconductor
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.3)
CRAN (R 4.0.0)
Bioconductor
CRAN (R 4.0.3)
Bioconductor
CRAN (R 4.0.3)
[1] /pstore/home/macnairw/lib/conda_r3.12
[2] /pstore/home/macnairw/.conda/envs/r_4.0.3/lib/R/library
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-conda-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /pstore/home/macnairw/.conda/envs/r_4.0.3/lib/libopenblasp-r0.3.12.so
locale:
[1] LC_CTYPE=C LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] grid parallel stats4 stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] scran_1.18.7 uwot_0.1.10
[3] scater_1.18.6 BiocParallel_1.24.1
[5] ggplot.multistats_1.0.0 seriation_1.2-9
[7] ComplexHeatmap_2.6.2 SeuratObject_4.0.0
[9] Seurat_4.0.1 conos_1.4.0
[11] igraph_1.2.6 SampleQC_0.4.5
[13] SingleCellExperiment_1.12.0 SummarizedExperiment_1.20.0
[15] Biobase_2.50.0 GenomicRanges_1.42.0
[17] GenomeInfoDb_1.26.7 IRanges_2.24.1
[19] S4Vectors_0.28.1 BiocGenerics_0.36.1
[21] MatrixGenerics_1.2.1 matrixStats_0.58.0
[23] Matrix_1.3-2 loomR_0.2.0
[25] itertools_0.1-3 iterators_1.0.13
[27] hdf5r_1.3.3 R6_2.5.0
[29] patchwork_1.1.1 forcats_0.5.1
[31] ggplot2_3.3.3 scales_1.1.1
[33] viridis_0.6.0 viridisLite_0.4.0
[35] assertthat_0.2.1 stringr_1.4.0
[37] data.table_1.14.0 magrittr_2.0.1
[39] circlize_0.4.12 RColorBrewer_1.1-2
[41] BiocStyle_2.18.1 colorout_1.2-2
[43] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] scattermore_0.7 tidyr_1.1.3
[3] bit64_4.0.5 knitr_1.32
[5] irlba_2.3.3 DelayedArray_0.16.3
[7] rpart_4.1-15 RCurl_1.98-1.3
[9] generics_0.1.0 callr_3.6.0
[11] mvnfast_0.2.5.1 cowplot_1.1.1
[13] usethis_2.0.1 RANN_2.6.1
[15] future_1.21.0 bit_4.0.4
[17] spatstat.data_2.1-0 httpuv_1.5.5
[19] xfun_0.22 jquerylib_0.1.3
[21] evaluate_0.14 promises_1.2.0.1
[23] TSP_1.1-10 fansi_0.4.2
[25] DBI_1.1.1 htmlwidgets_1.5.3
[27] spatstat.geom_2.1-0 purrr_0.3.4
[29] ellipsis_0.3.1 dplyr_1.0.5
[31] deldir_0.2-10 sparseMatrixStats_1.2.1
[33] vctrs_0.3.7 remotes_2.3.0
[35] Cairo_1.5-12.2 ROCR_1.0-11
[37] abind_1.4-5 cachem_1.0.4
[39] withr_2.4.2 grr_0.9.5
[41] sctransform_0.3.2 prettyunits_1.1.1
[43] mclust_5.4.7 goftest_1.2-2
[45] cluster_2.1.2 segmented_1.3-3
[47] lazyeval_0.2.2 crayon_1.4.1
[49] edgeR_3.32.1 pkgconfig_2.0.3
[51] labeling_0.4.2 pkgload_1.2.1
[53] nlme_3.1-152 vipor_0.4.5
[55] devtools_2.4.0 rlang_0.4.10
[57] globals_0.14.0 lifecycle_1.0.0
[59] miniUI_0.1.1.1 registry_0.5-1
[61] rsvd_1.0.5 rprojroot_2.0.2
[63] polyclip_1.10-0 lmtest_0.9-38
[65] zoo_1.8-9 Matrix.utils_0.9.8
[67] beeswarm_0.3.1 processx_3.5.1
[69] whisker_0.4 ggridges_0.5.3
[71] GlobalOptions_0.1.2 png_0.1-7
[73] rjson_0.2.20 bitops_1.0-6
[75] KernSmooth_2.23-18 DelayedMatrixStats_1.12.3
[77] shape_1.4.5 parallelly_1.24.0
[79] sccore_0.1.2 beachmat_2.6.4
[81] memoise_2.0.0 plyr_1.8.6
[83] hexbin_1.28.2 ica_1.0-2
[85] zlibbioc_1.36.0 compiler_4.0.3
[87] dqrng_0.2.1 clue_0.3-59
[89] fitdistrplus_1.1-3 cli_2.4.0
[91] XVector_0.30.0 listenv_0.8.0
[93] ps_1.6.0 pbapply_1.4-3
[95] MASS_7.3-53.1 mgcv_1.8-35
[97] tidyselect_1.1.0 stringi_1.5.3
[99] highr_0.9 yaml_2.2.1
[101] BiocSingular_1.6.0 locfit_1.5-9.4
[103] ggrepel_0.9.1 sass_0.3.1
[105] tools_4.0.3 future.apply_1.7.0
[107] rstudioapi_0.13 bluster_1.0.0
[109] foreach_1.5.1 git2r_0.28.0
[111] gridExtra_2.3 farver_2.1.0
[113] Rtsne_0.15 digest_0.6.27
[115] BiocManager_1.30.12 shiny_1.6.0
[117] Rcpp_1.0.6 scuttle_1.0.4
[119] later_1.1.0.1 RcppAnnoy_0.0.18
[121] httr_1.4.2 kernlab_0.9-29
[123] colorspace_2.0-0 fs_1.5.0
[125] tensor_1.5 reticulate_1.18
[127] splines_4.0.3 statmod_1.4.35
[129] spatstat.utils_2.1-0 sessioninfo_1.1.1
[131] plotly_4.9.3 xtable_1.8-4
[133] jsonlite_1.7.2 leidenAlg_0.1.1
[135] testthat_3.0.2 pillar_1.6.0
[137] htmltools_0.5.1.1 mime_0.10
[139] glue_1.4.2 fastmap_1.1.0
[141] BiocNeighbors_1.8.2 codetools_0.2-18
[143] pkgbuild_1.2.0 mvtnorm_1.1-1
[145] utf8_1.2.1 lattice_0.20-41
[147] bslib_0.2.4 spatstat.sparse_2.0-0
[149] tibble_3.1.1 mixtools_1.2.0
[151] ggbeeswarm_0.6.0 leiden_0.3.7
[153] gtools_3.8.2 survival_3.2-10
[155] limma_3.46.0 rmarkdown_2.7
[157] desc_1.3.0 munsell_0.5.0
[159] GetoptLong_1.0.5 GenomeInfoDbData_1.2.4
[161] reshape2_1.4.4 gtable_0.3.0
[163] spatstat.core_2.1-2