Figures for manuscript
Will Macnair
Institute for Molecular Life Sciences, University of Zurich, SwitzerlandSwiss Institute of Bioinformatics (SIB), University of Zurich, SwitzerlandDecember 20, 2021
Last updated: 2021-12-20
Checks: 4 3
Knit directory: MS_lesions/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
The global environment had objects present when the code in the R Markdown file was run. These objects can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment. Use wflow_publish or wflow_build to ensure that the code is always run in an empty environment.
The following objects were defined in the global environment when these results were created:
| Name | Class | Size |
|---|---|---|
| q | function | 1008 bytes |
The command set.seed(20210118) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
- ancom_bootstrapped_gm_4pcs
- ancom_bootstrapped_gm
- ancom_bootstrapped_wm
- dotplot_marker_genes
- effect_of_including_pcs_neurons
- effect_of_including_pcs_others
- fixed_vs_random_gm_4pcs
- fixed_vs_random_gm
- fixed_vs_random_wm
- gm_vs_wm_proportions
- grp17_cell_abundances
- module_go_terms_annotations
- module_scores_opc_oligo
- oligo_barplot_gm
- oligo_barplot_wm
- plot_dotplot_dheeraj_compact
- plot_test
- plot_umap_all_celltypes
- plot_umap_final_celltypes_sel
- plot_umap_opc_oligo_only
- save_outputs
- session_info
- session-info-chunk-inserted-by-workflowr
- umap_all_celltypes
- umap_opc_oligo
To ensure reproducibility of the results, delete the cache directory ms00_manuscript_figures_cache and re-run the analysis. To have workflowr automatically delete the cache directory prior to building the file, set delete_cache = TRUE when running wflow_build() or wflow_publish().
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 1a38d84. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rprofile
Ignored: .Rproj.user/
Ignored: ._MS_lesions.sublime-project
Ignored: .log/
Ignored: MS_lesions.sublime-project
Ignored: MS_lesions.sublime-workspace
Ignored: analysis/.__site.yml
Ignored: analysis/fig_muscat_cache/
Ignored: analysis/ms00_manuscript_figures_cache/
Ignored: analysis/ms02_doublet_id_cache/
Ignored: analysis/ms03_SampleQC_cache/
Ignored: analysis/ms04_conos_cache/
Ignored: analysis/ms05_splitting_cache/
Ignored: analysis/ms06_sccaf_cache/
Ignored: analysis/ms07_soup_cache/
Ignored: analysis/ms08_modules_cache/
Ignored: analysis/ms08_modules_pseudobulk_cache/
Ignored: analysis/ms09_ancombc_cache/
Ignored: analysis/ms09_ancombc_clean_1e3_cache/
Ignored: analysis/ms09_ancombc_clean_2e3_cache/
Ignored: analysis/ms09_ancombc_mixed_cache/
Ignored: analysis/ms10_muscat_run01_cache/
Ignored: analysis/ms10_muscat_run02_cache/
Ignored: analysis/ms10_muscat_template_broad_slim_cache/
Ignored: analysis/ms10_muscat_template_fine_slim_cache/
Ignored: analysis/ms11_paga_cache/
Ignored: analysis/ms12_markers_cache/
Ignored: analysis/ms13_labelling_cache/
Ignored: analysis/ms14_lesions_cache/
Ignored: analysis/ms15_mofa_sample_gm_cache/
Ignored: analysis/ms15_mofa_sample_gm_final_meta_cache/
Ignored: analysis/ms15_mofa_sample_gm_superclean_cache/
Ignored: analysis/ms15_mofa_sample_gm_w_layers_final_meta_cache/
Ignored: analysis/ms15_mofa_sample_wm_cache/
Ignored: analysis/ms15_mofa_sample_wm_final_meta_cache/
Ignored: analysis/ms15_mofa_sample_wm_new_meta_cache/
Ignored: analysis/ms15_mofa_sample_wm_superclean_cache/
Ignored: analysis/ms15_patients_cache/
Ignored: analysis/ms15_patients_gm_cache/
Ignored: analysis/ms15_patients_sample_level_cache/
Ignored: analysis/ms15_patients_w_ms_cache/
Ignored: analysis/supp06_sccaf_cache/
Ignored: analysis/supp07_superclean_check_cache/
Ignored: analysis/supp09_ancombc_cache/
Ignored: analysis/supp09_ancombc_mixed_cache/
Ignored: analysis/supp09_ancombc_rowitch_cache/
Ignored: analysis/supp09_ancombc_superclean_cache/
Ignored: analysis/supp10_muscat_cache/
Ignored: analysis/supp10_muscat_ctrl_gm_vs_wm_cache/
Ignored: analysis/supp10_muscat_gm_layers_effects_cache/
Ignored: analysis/supp10_muscat_gsea_cache/
Ignored: analysis/supp10_muscat_heatmaps_cache/
Ignored: analysis/supp10_muscat_olg_pc1_cache/
Ignored: analysis/supp10_muscat_olg_pc2_cache/
Ignored: analysis/supp10_muscat_olg_pc_cache/
Ignored: analysis/supp10_muscat_regression_cache/
Ignored: analysis/supp10_muscat_soup_cache/
Ignored: analysis/supp10_muscat_soup_mito_cache/
Ignored: code/._ms10_muscat_fns_recover.R
Ignored: code/.recovery/
Ignored: code/jobs/._muscat_run09_2021-10-11.slurm
Ignored: code/muscat_plan.txt
Ignored: data/
Ignored: figures/
Ignored: output/
Ignored: tmp/
Untracked files:
Untracked: Rplots.pdf
Untracked: analysis/supp09_ancombc_superclean.Rmd
Untracked: code/dev_dheeraj_oligo_dotplots_2021-12-08.R
Untracked: code/dev_magma_de_heatmaps_2021-12-03.R
Untracked: code/dev_plot_fcs_selected_genes_2021-12-09.R
Untracked: code/jobs/muscat_run20_2021-12-07.slurm
Unstaged changes:
Modified: analysis/fig_muscat.Rmd
Modified: analysis/ms00_manuscript_figures.Rmd
Modified: analysis/ms09_ancombc_mixed.Rmd
Modified: analysis/ms13_labelling.Rmd
Modified: analysis/ms14_lesions.Rmd
Modified: analysis/ms15_mofa_sample_wm_superclean.Rmd
Modified: analysis/supp07_superclean_check.Rmd
Modified: code/dev_de_w_contamation_2021-10-25.R
Modified: code/dev_edger_on_mofa_20210804.R
Modified: code/fig_muscat.R
Modified: code/ms04_conos.R
Modified: code/ms09_ancombc_mixed.R
Modified: code/ms14_lesions.R
Modified: code/supp10_muscat.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ms00_manuscript_figures.Rmd) and HTML (public/ms00_manuscript_figures.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 1a38d84 | wmacnair | 2021-12-17 | Add initial version of file with all figures |
| html | 1a38d84 | wmacnair | 2021-12-17 | Add initial version of file with all figures |
Setup / definitions
Libraries
Helper functions
source('code/ms00_utils.R')
library("knitr")Figure 1
B-1
UMAP of all celltypes
include_graphics("figure/ms12_markers.Rmd/plot_umap_final_celltypes_sel-1.png", error = FALSE)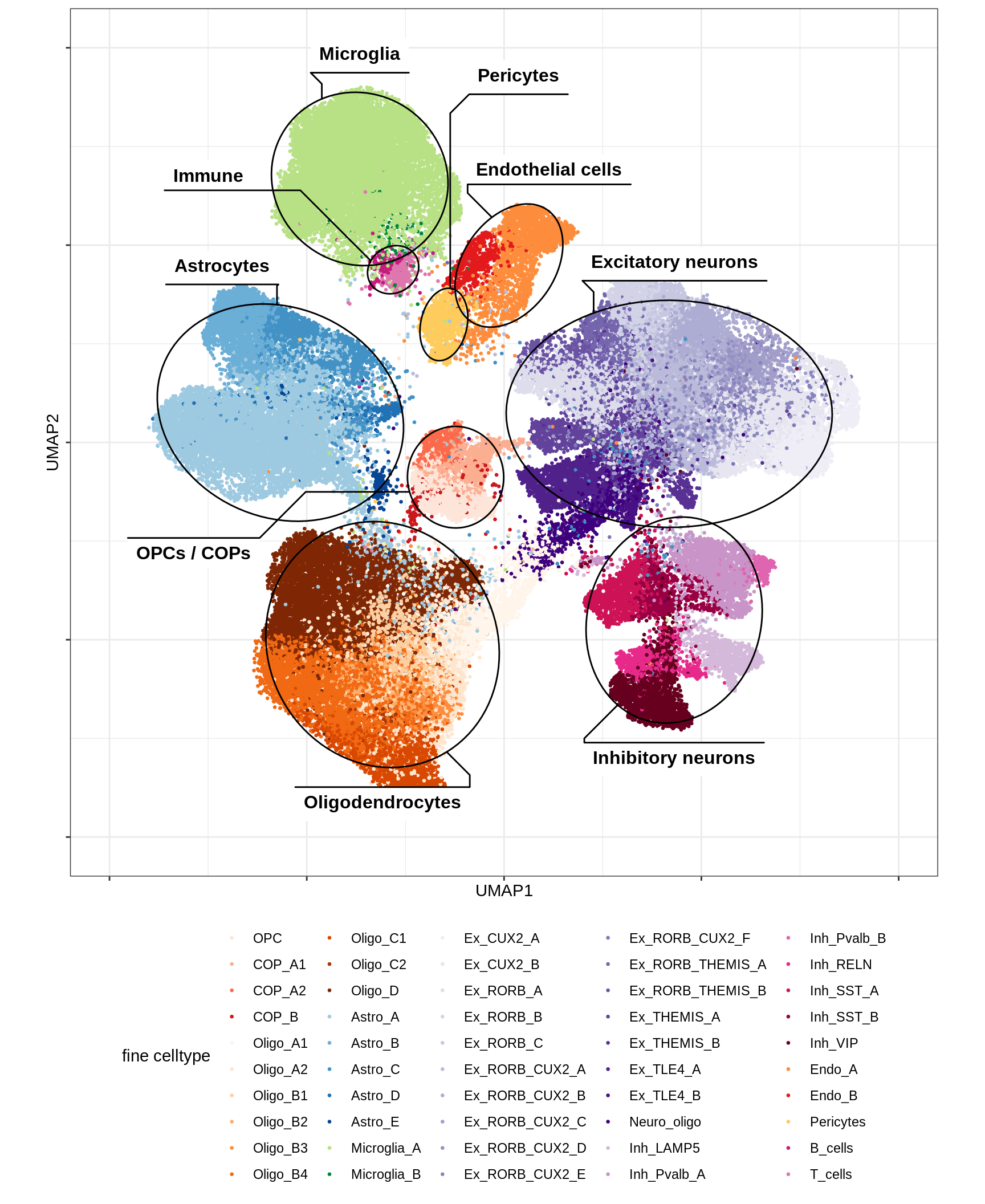
| Version | Author | Date |
|---|---|---|
| 7fb1b95 | wmacnair | 2021-11-25 |
B-2
Dotplot of selected marker genes
include_graphics("figure/ms12_markers.Rmd/plot_dotplot_dheeraj_compact-1.png", error = FALSE)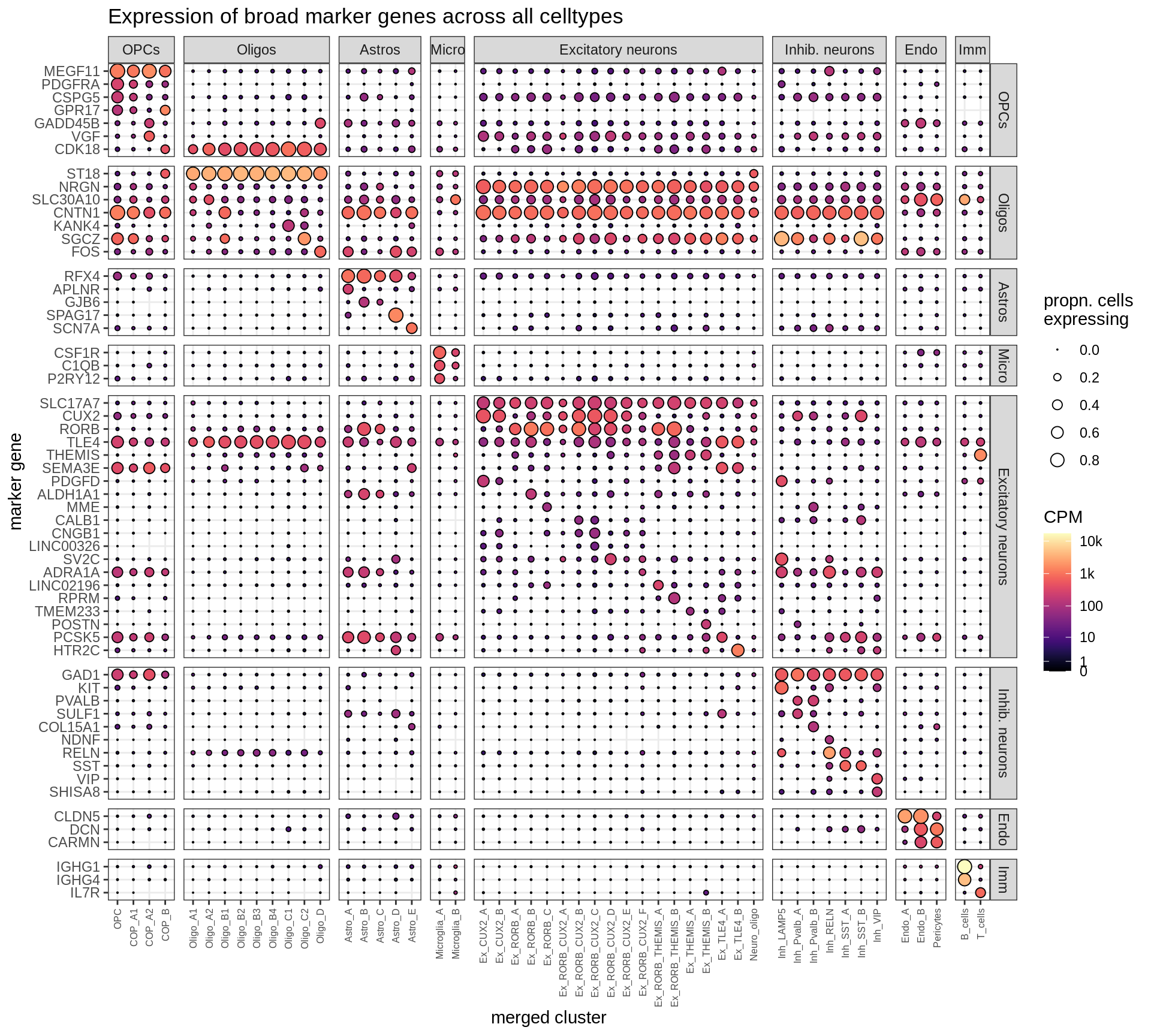
| Version | Author | Date |
|---|---|---|
| 7fb1b95 | wmacnair | 2021-11-25 |
C-1
UMAP of just OPCs and oligos
include_graphics("figure/ms12_markers.Rmd/plot_umap_opc_oligo_only-1.png", error = FALSE)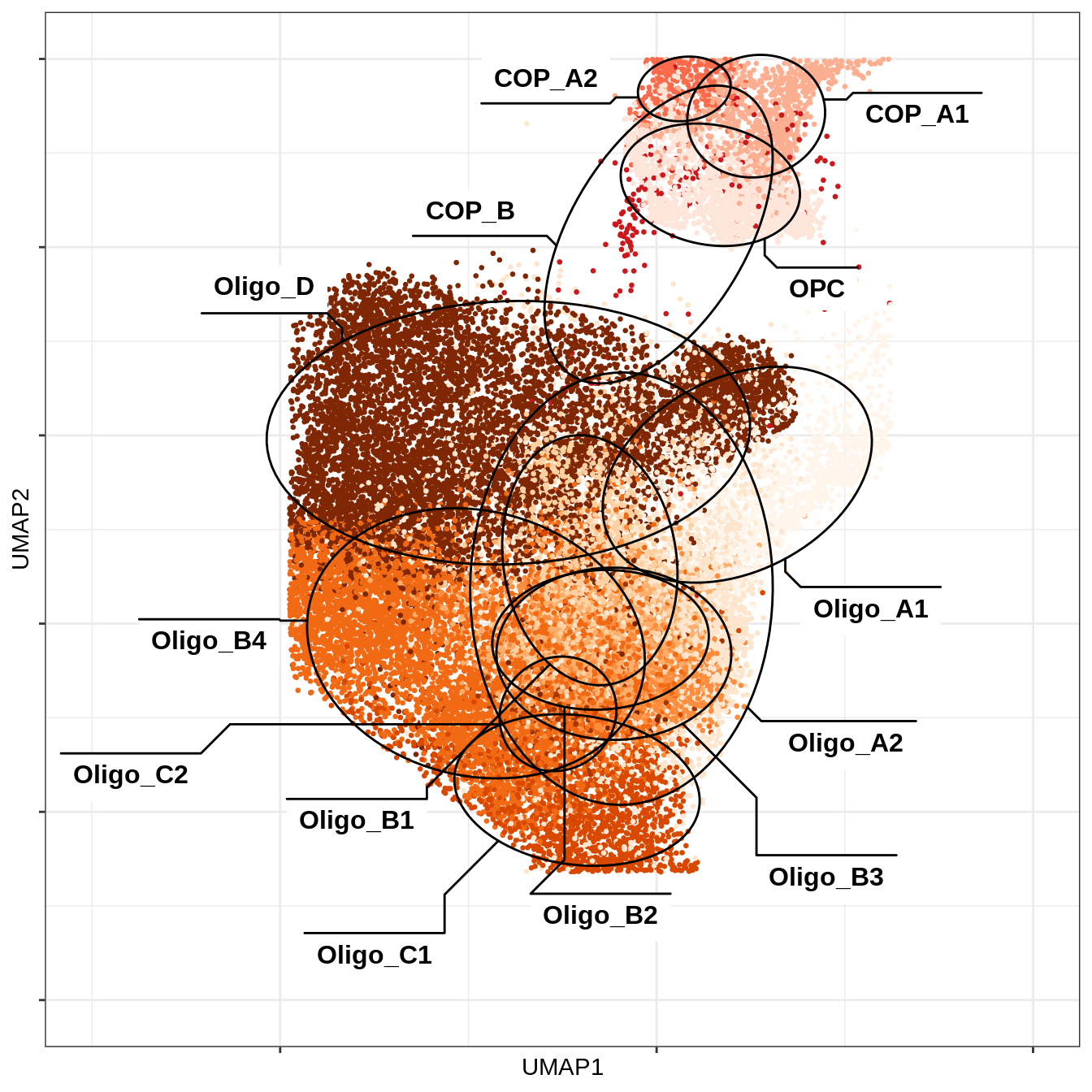
| Version | Author | Date |
|---|---|---|
| 7fb1b95 | wmacnair | 2021-11-25 |
C-2
Module scores for OPCs and oligos
include_graphics("figure/ms08_modules.Rmd/plot_scores_by_type_scaled-1.png", error = FALSE)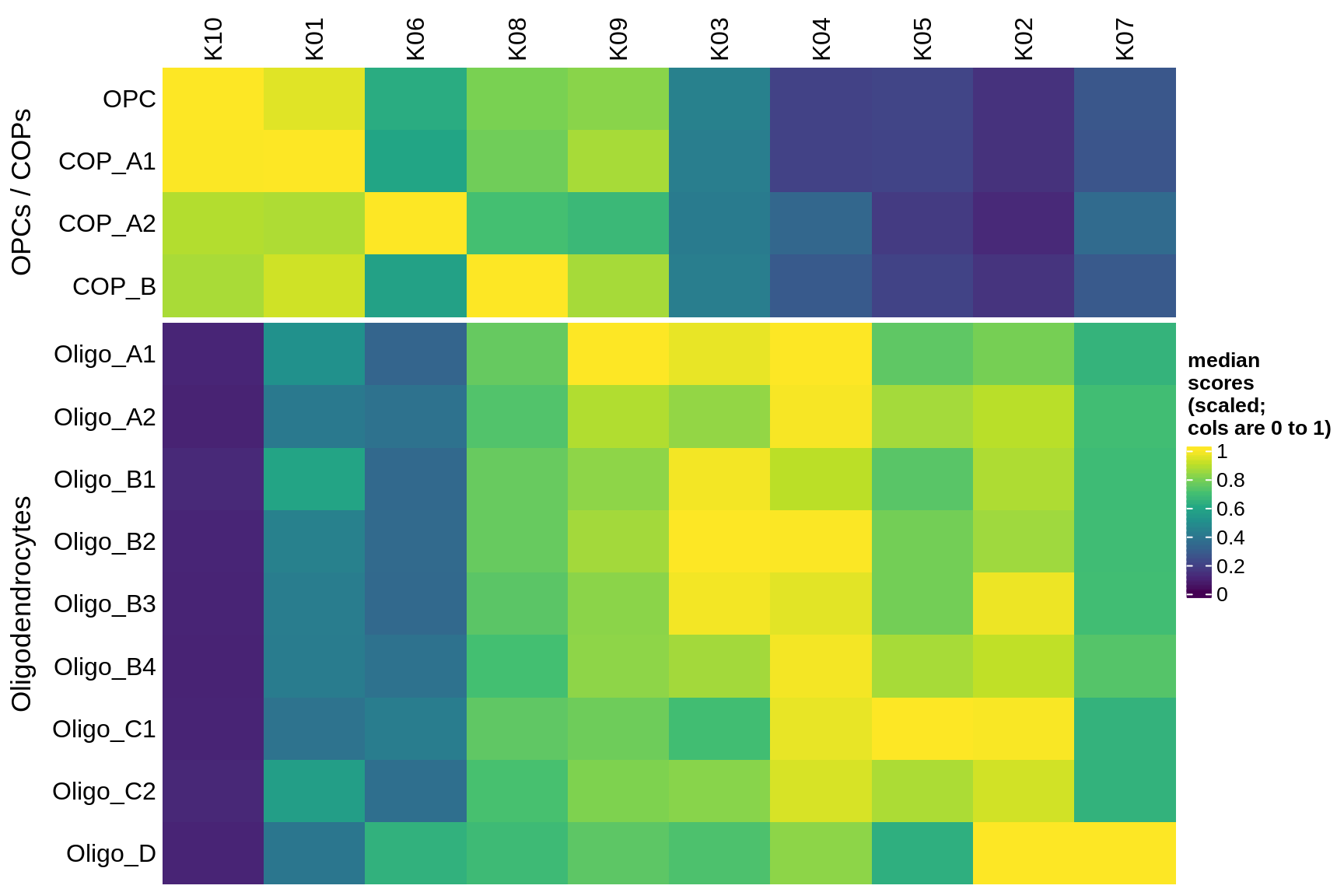
C-3
GO term annotations for OPC and oligo modules
include_graphics("figure/ms08_modules.Rmd/plot_annotations_std-1.png", error = FALSE)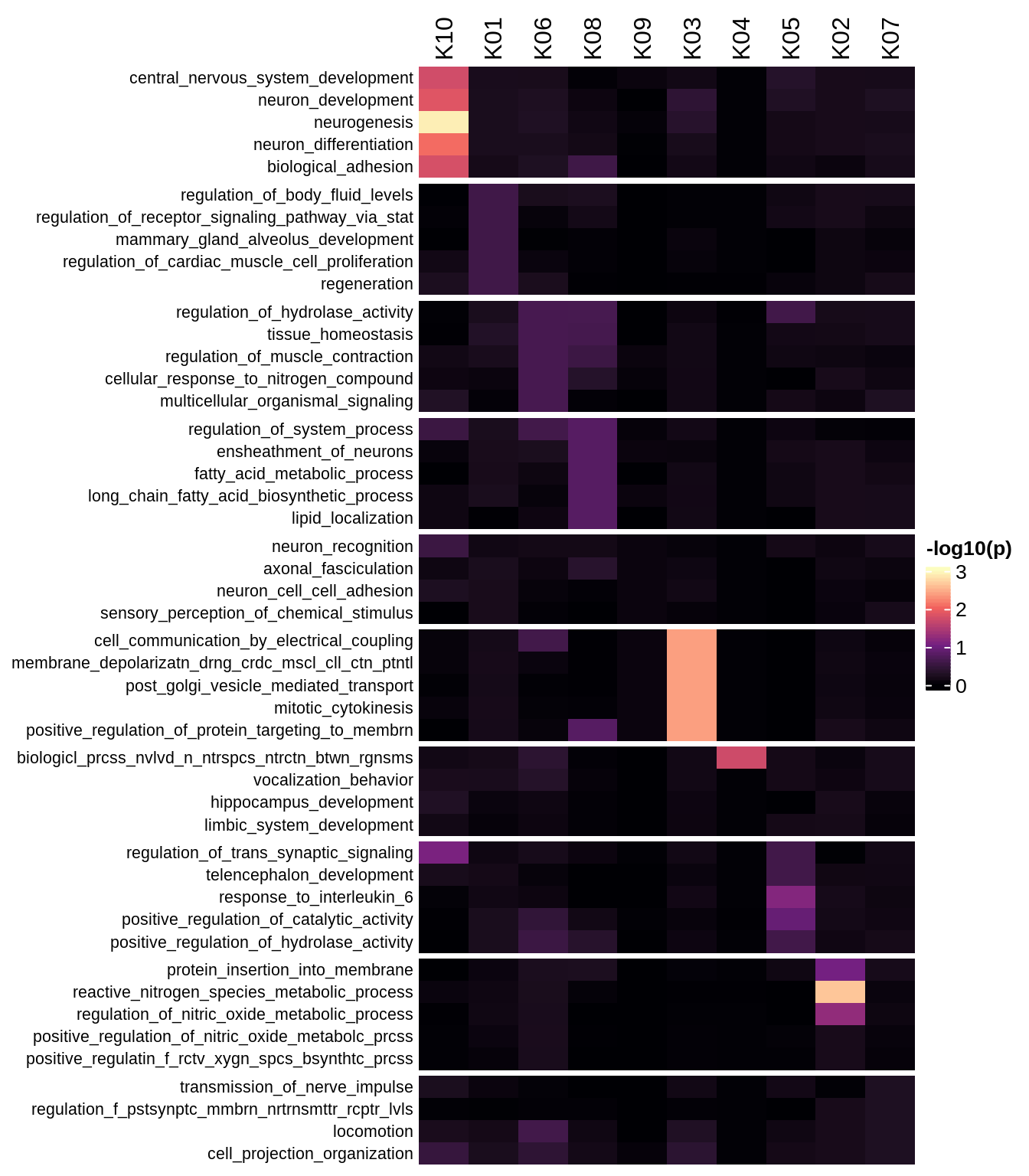
Figure 2
A
Contribution to variability in celltype abundances explained by lesion + patient in WM
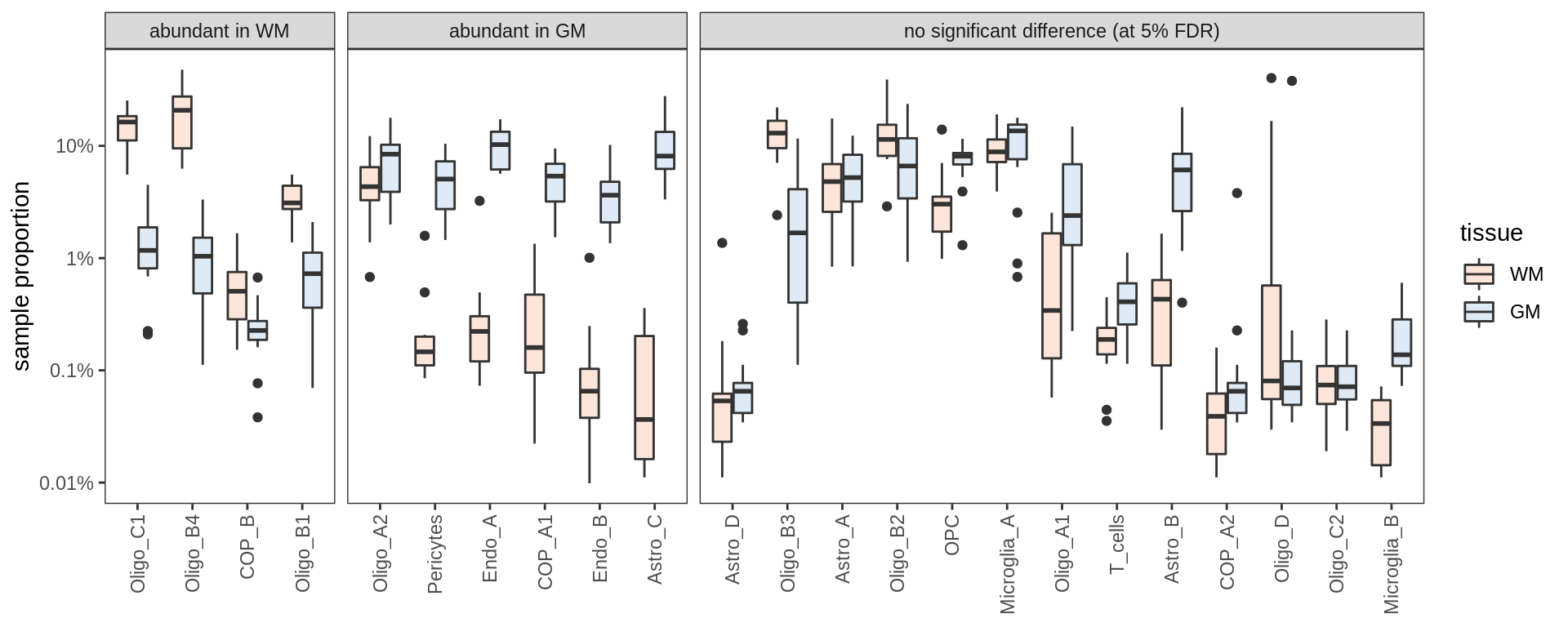
include_graphics("figure/ms09_ancombc_mixed.Rmd/plot_lrt_results-1.png", error = FALSE)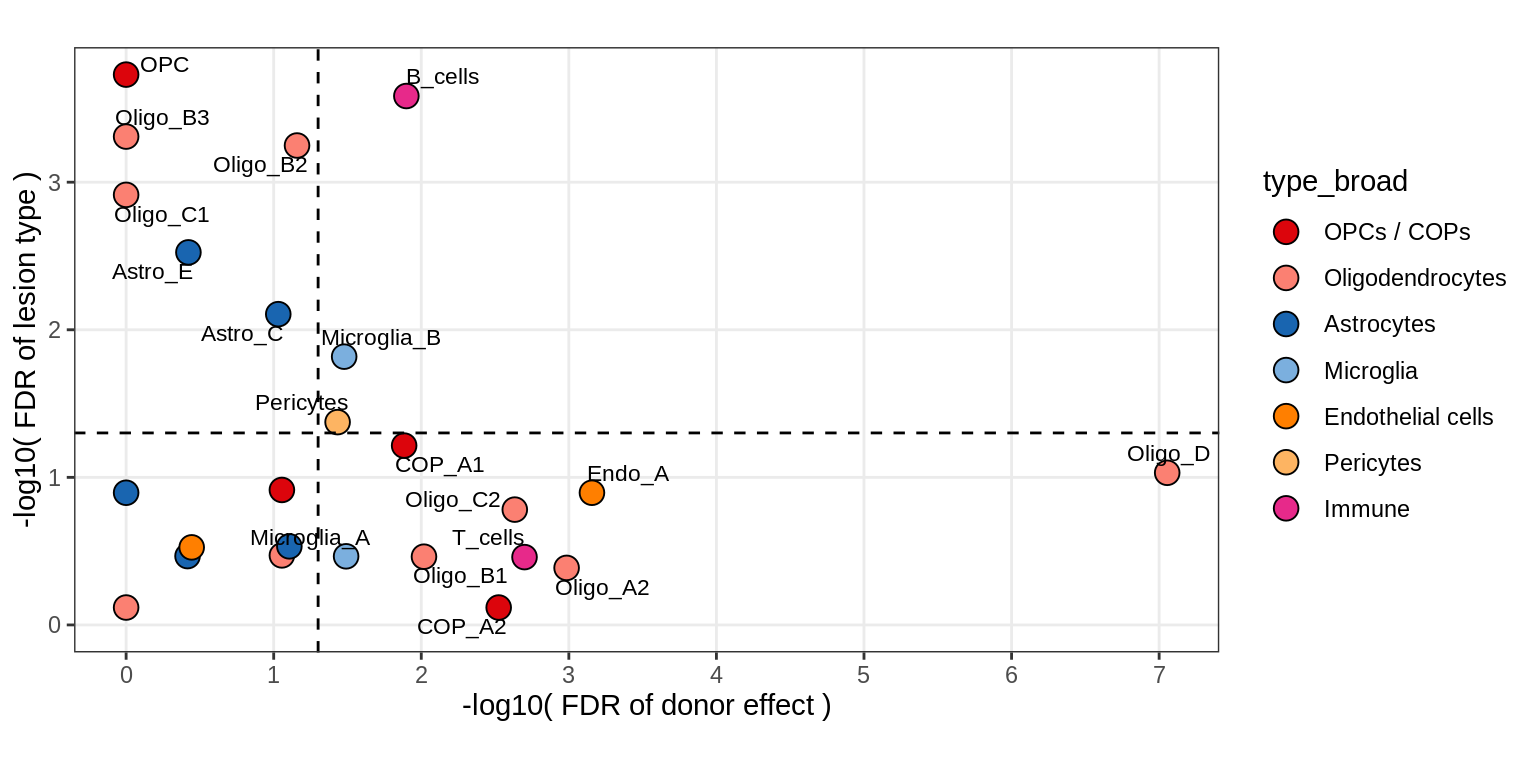
B
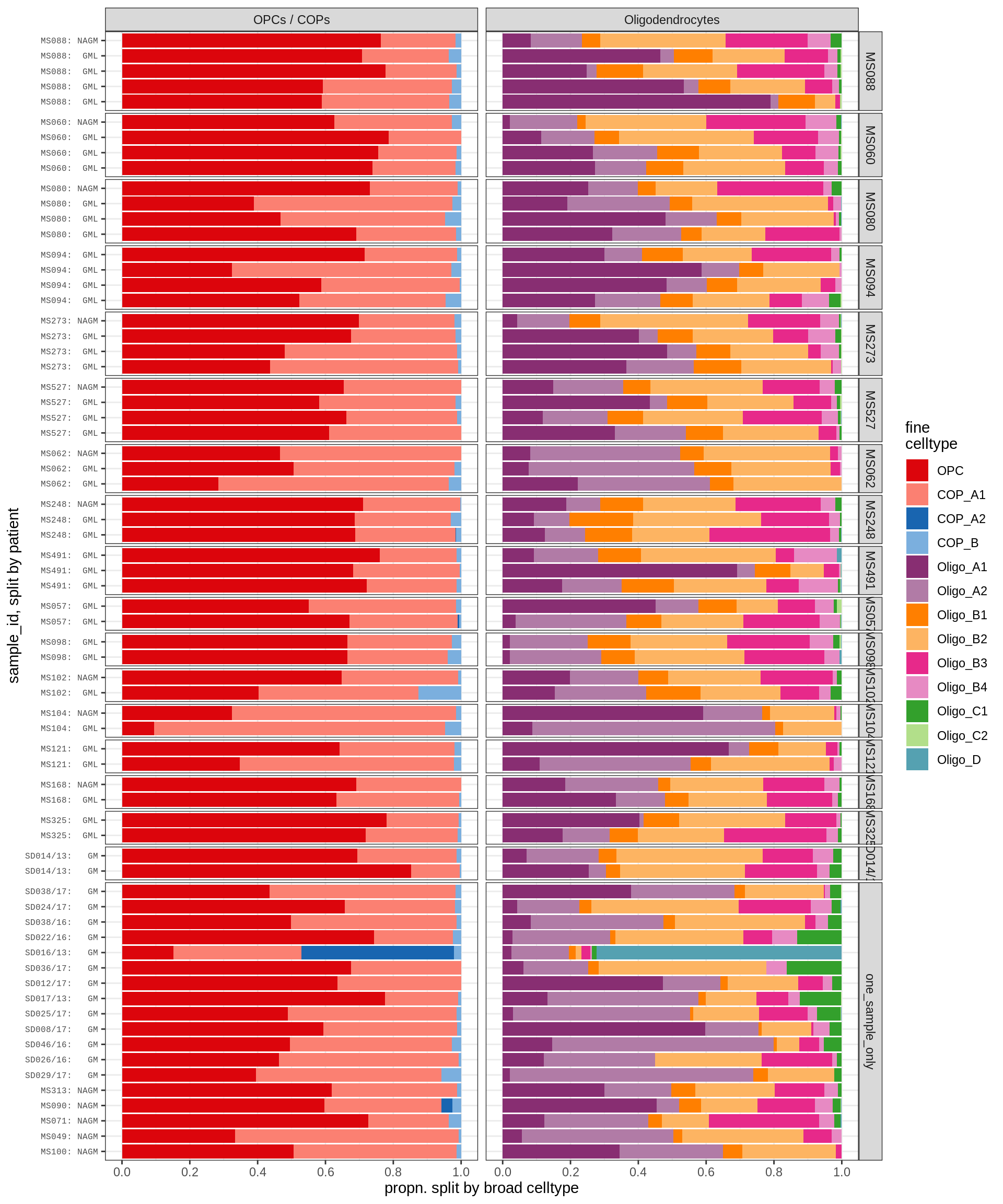
WM oligodendroglia proportions barplot
include_graphics("figure/ms09_ancombc_mixed.Rmd/plot_sample_splits_bars_oligos-1.png", error = FALSE)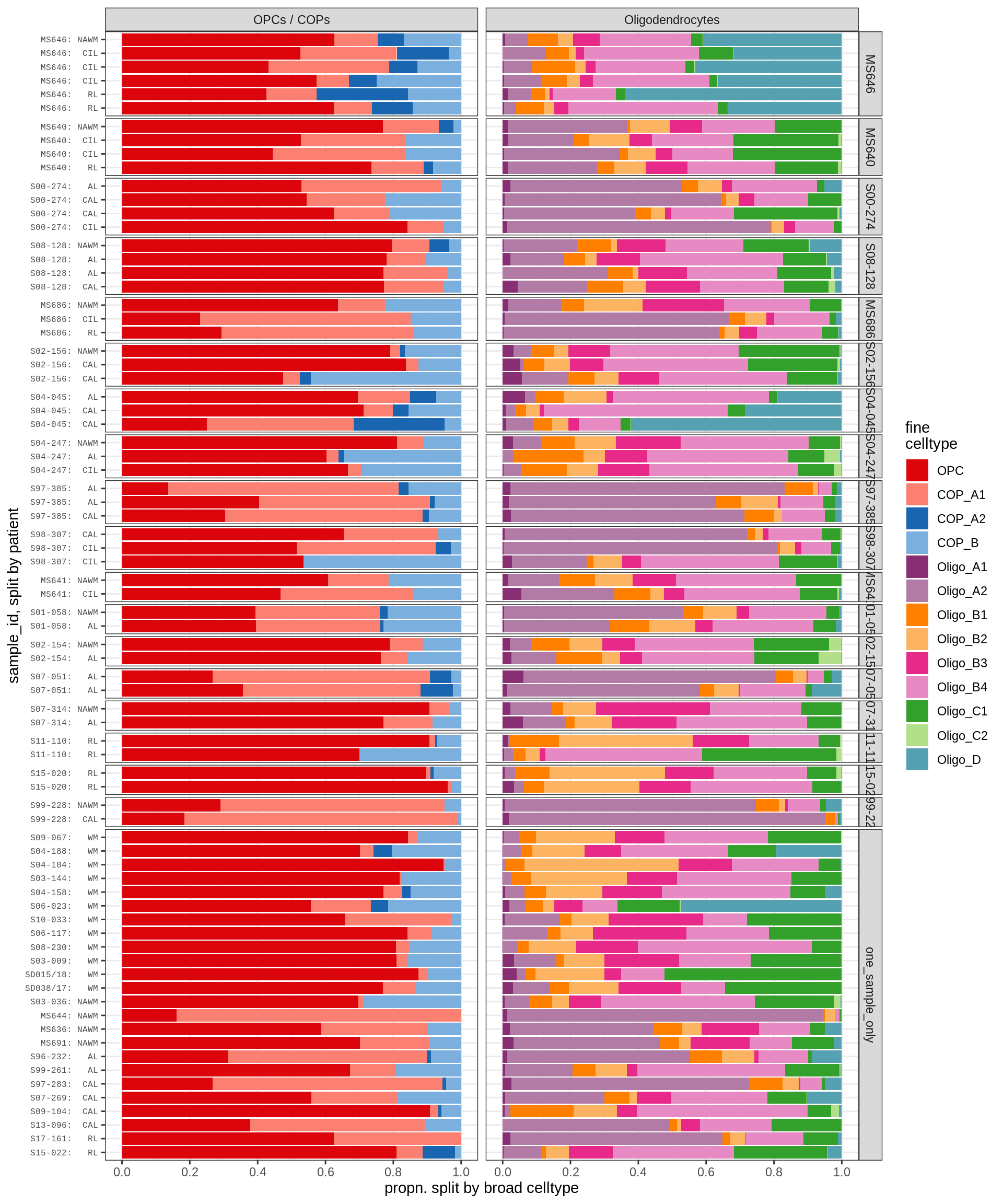
C
Differential abundance results for WM
include_graphics("figure/ms09_ancombc_mixed.Rmd/plot_bootstraps_lesions-1.png", error = FALSE)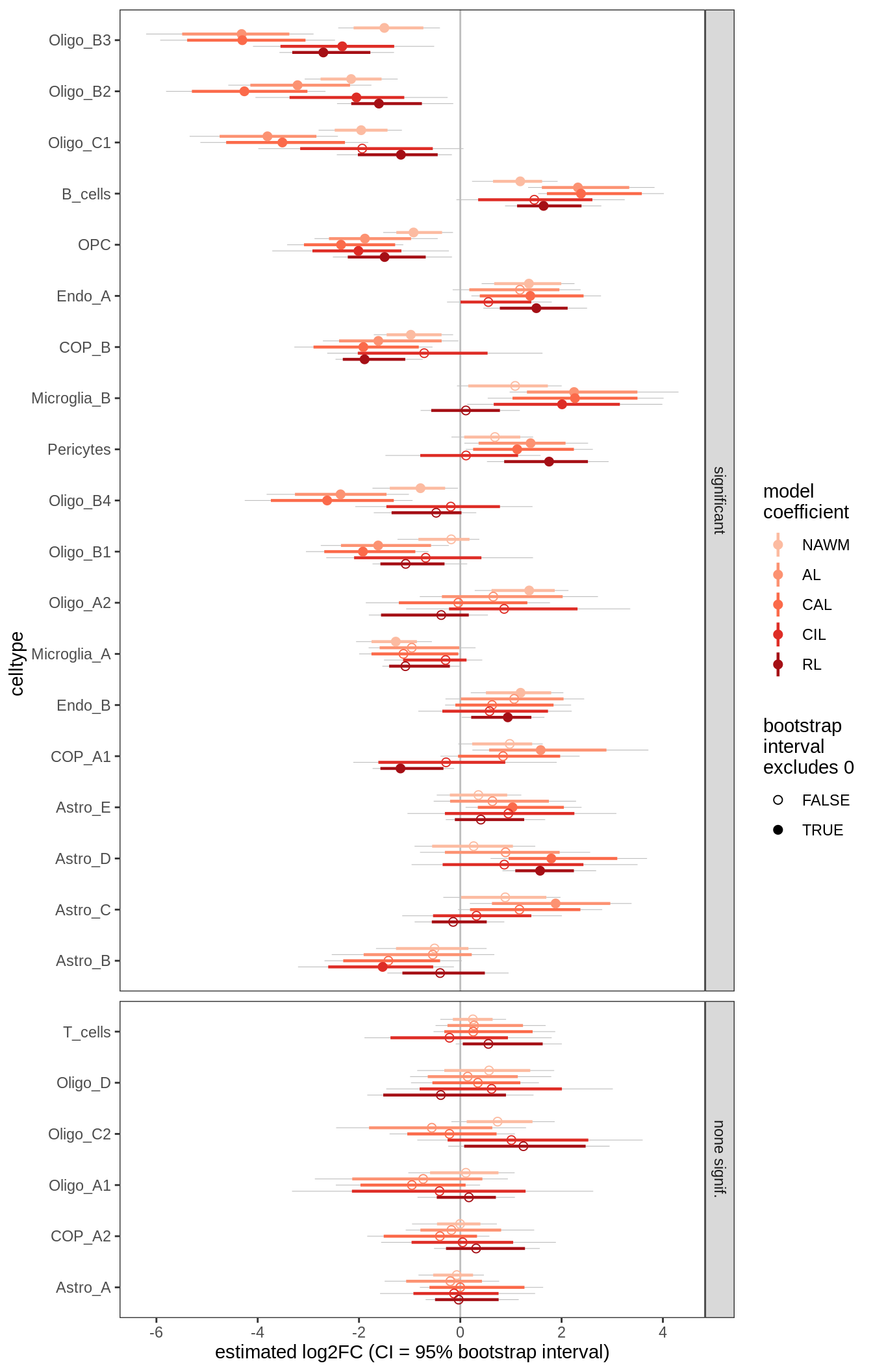
D
Comparison of Sarah’s validation of GPR17-expressing cells
include_graphics("figure/ms09_ancombc_mixed.Rmd/plot_no_gpr17_cells-1.png", error = FALSE)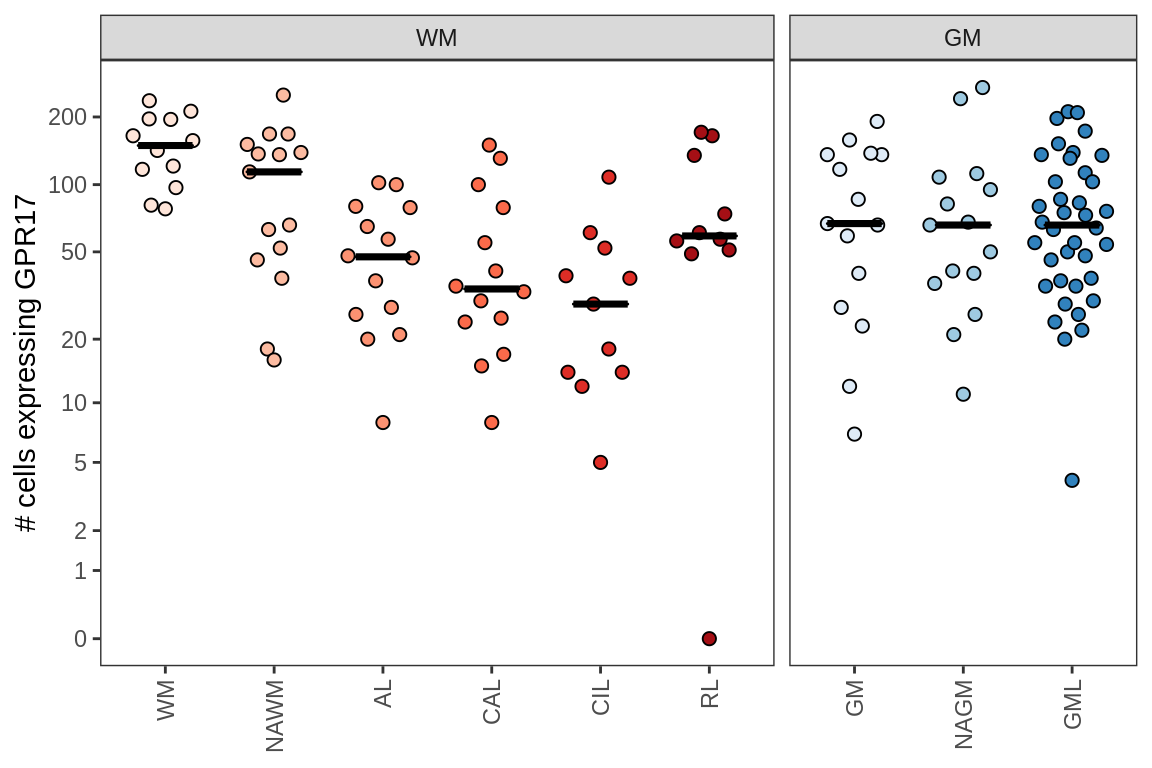
Figure 4
A
Illustration of random effects model
include_graphics("figure/ms15_mofa_sample_wm_final_meta.Rmd/fig_random_effects_example-3.png", error = FALSE)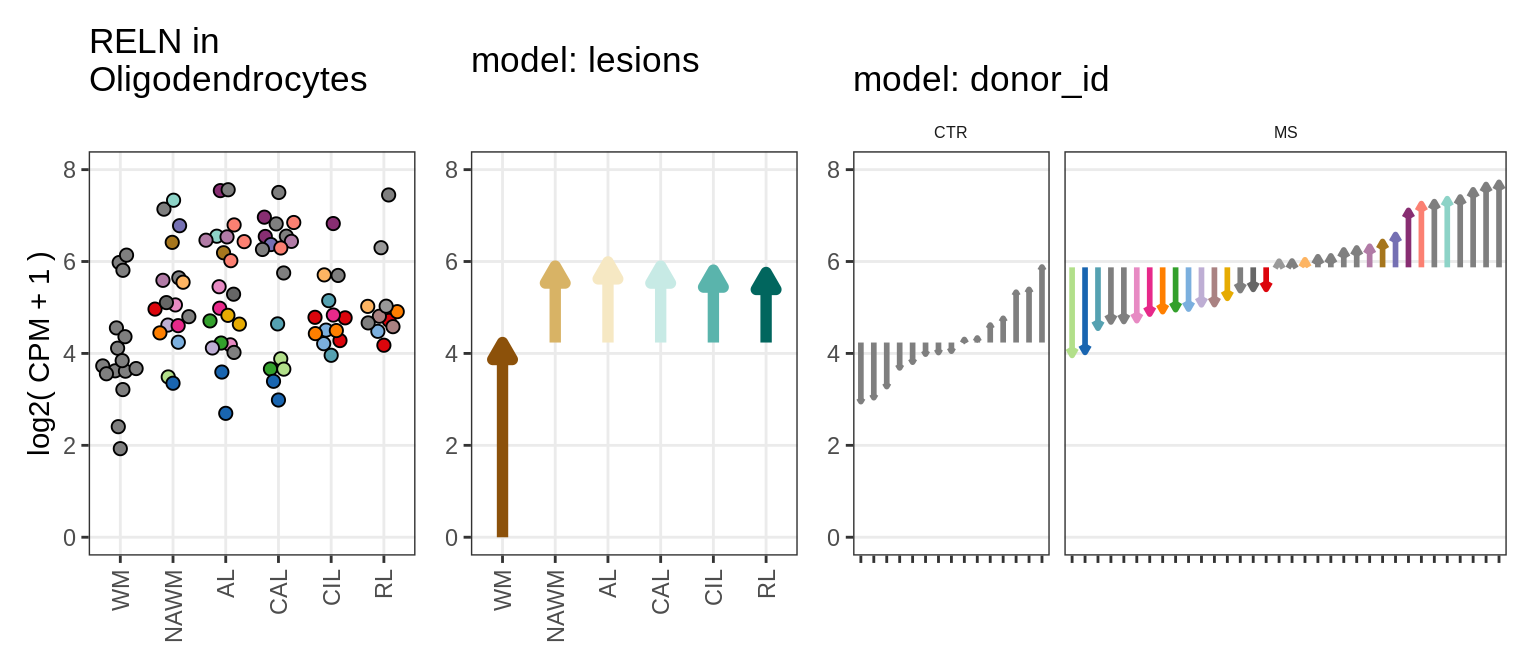
| Version | Author | Date |
|---|---|---|
| 7fb1b95 | wmacnair | 2021-11-25 |
B
Disease effect vs donor effect
include_graphics("figure/ms15_mofa_sample_wm_final_meta.Rmd/plot_muscat_vs_sd_min-3.png", error = FALSE)
C
MOFA factors
include_graphics("figure/ms15_mofa_sample_wm_final_meta.Rmd/fig_mofa_factors_diagnosis-1.png", error = FALSE)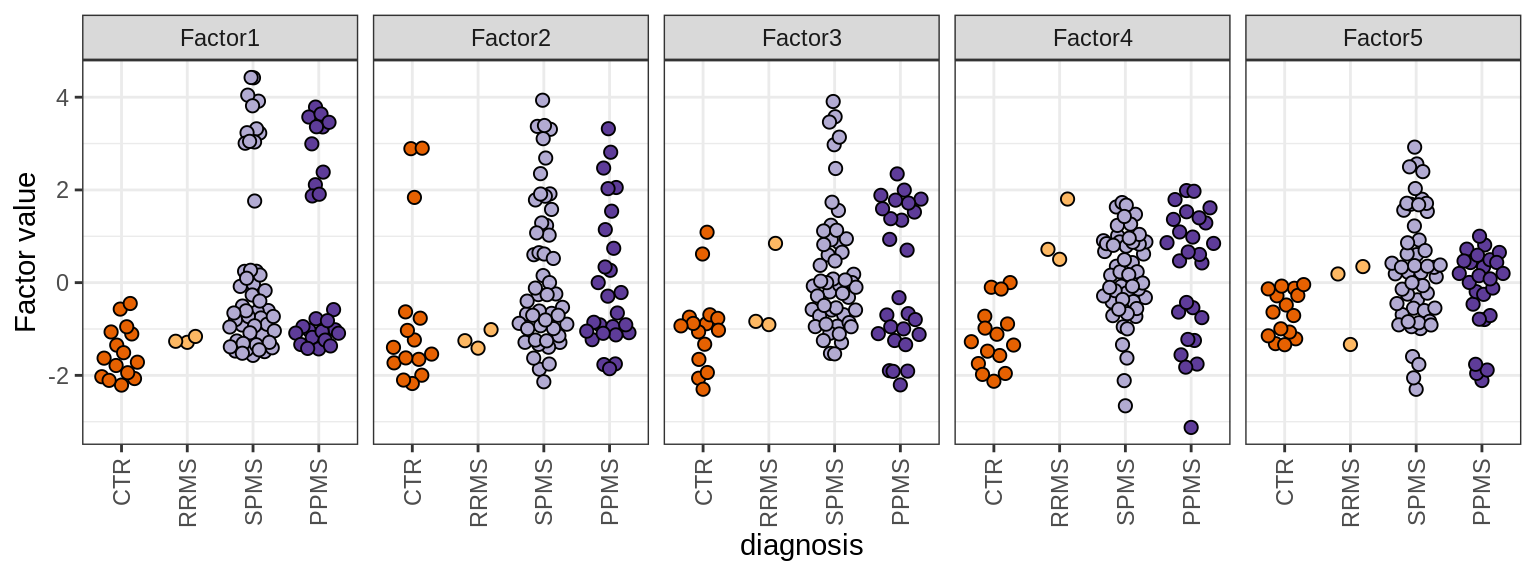
| Version | Author | Date |
|---|---|---|
| 7fb1b95 | wmacnair | 2021-11-25 |
D
Variance explained
include_graphics("figure/ms15_mofa_sample_wm_final_meta.Rmd/fig_factor_r2s-1.png", error = FALSE)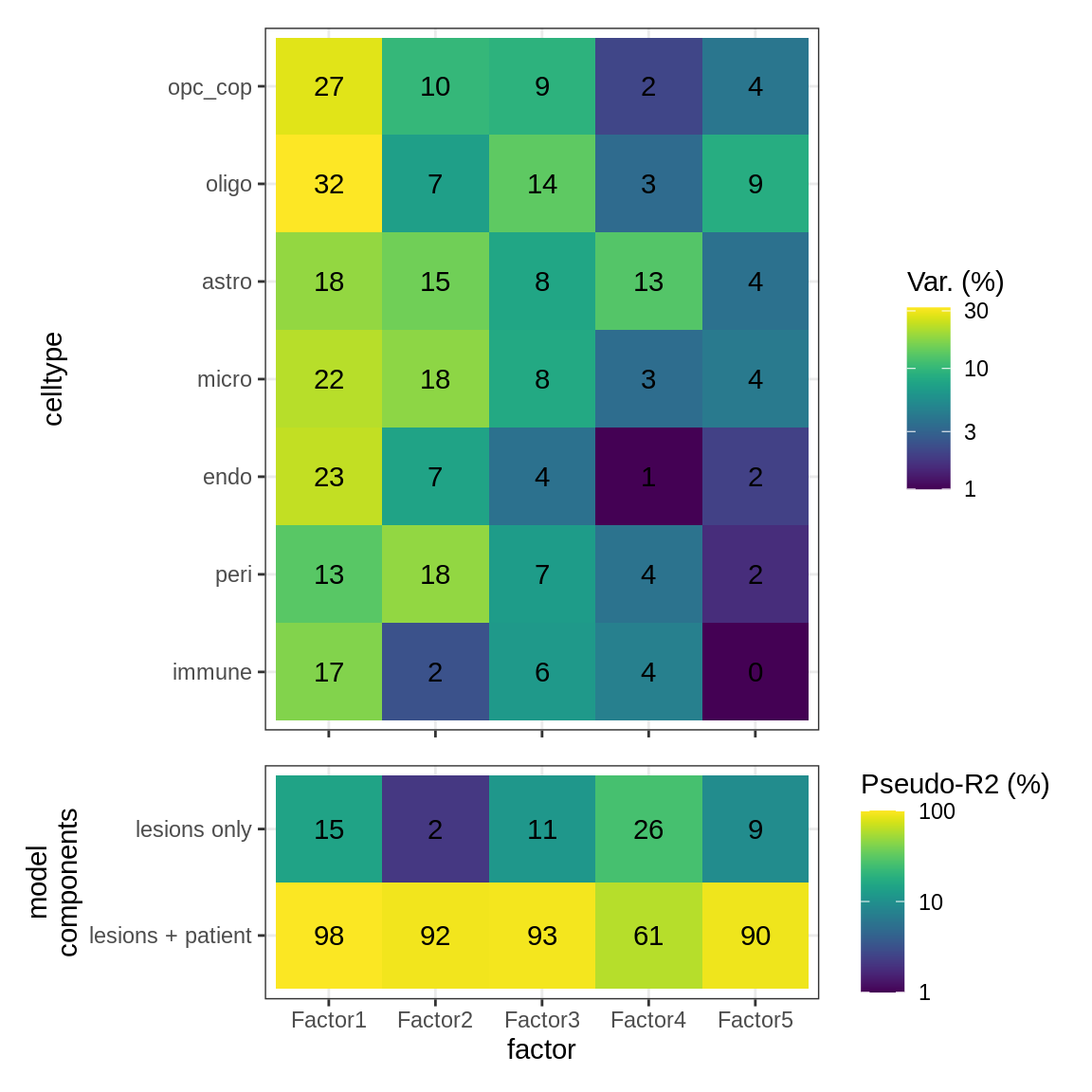
| Version | Author | Date |
|---|---|---|
| 7fb1b95 | wmacnair | 2021-11-25 |
E
Patient stratification
include_graphics("figure/ms15_mofa_sample_wm_final_meta.Rmd/fig_f1_vs_f2-3.png", error = FALSE)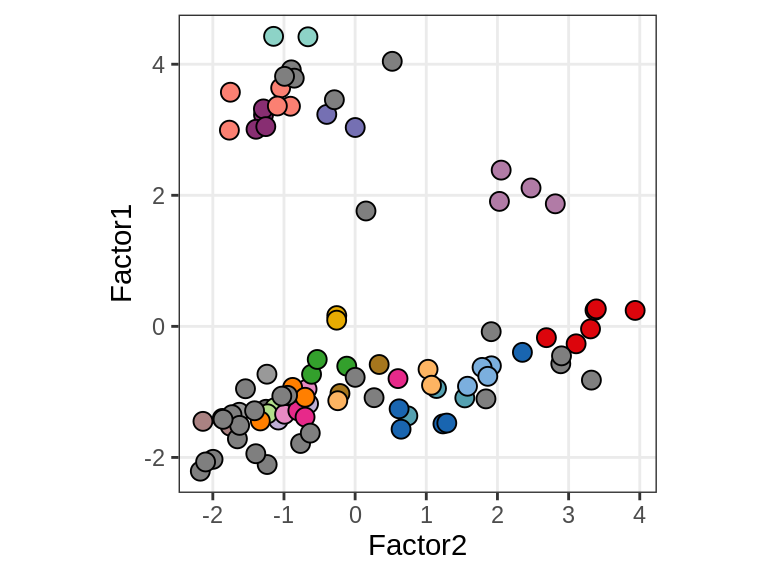
| Version | Author | Date |
|---|---|---|
| 7fb1b95 | wmacnair | 2021-11-25 |
F
Factor 1 top genes
include_graphics("figure/ms15_mofa_sample_wm_final_meta.Rmd/fig_factor1-1.png", error = FALSE)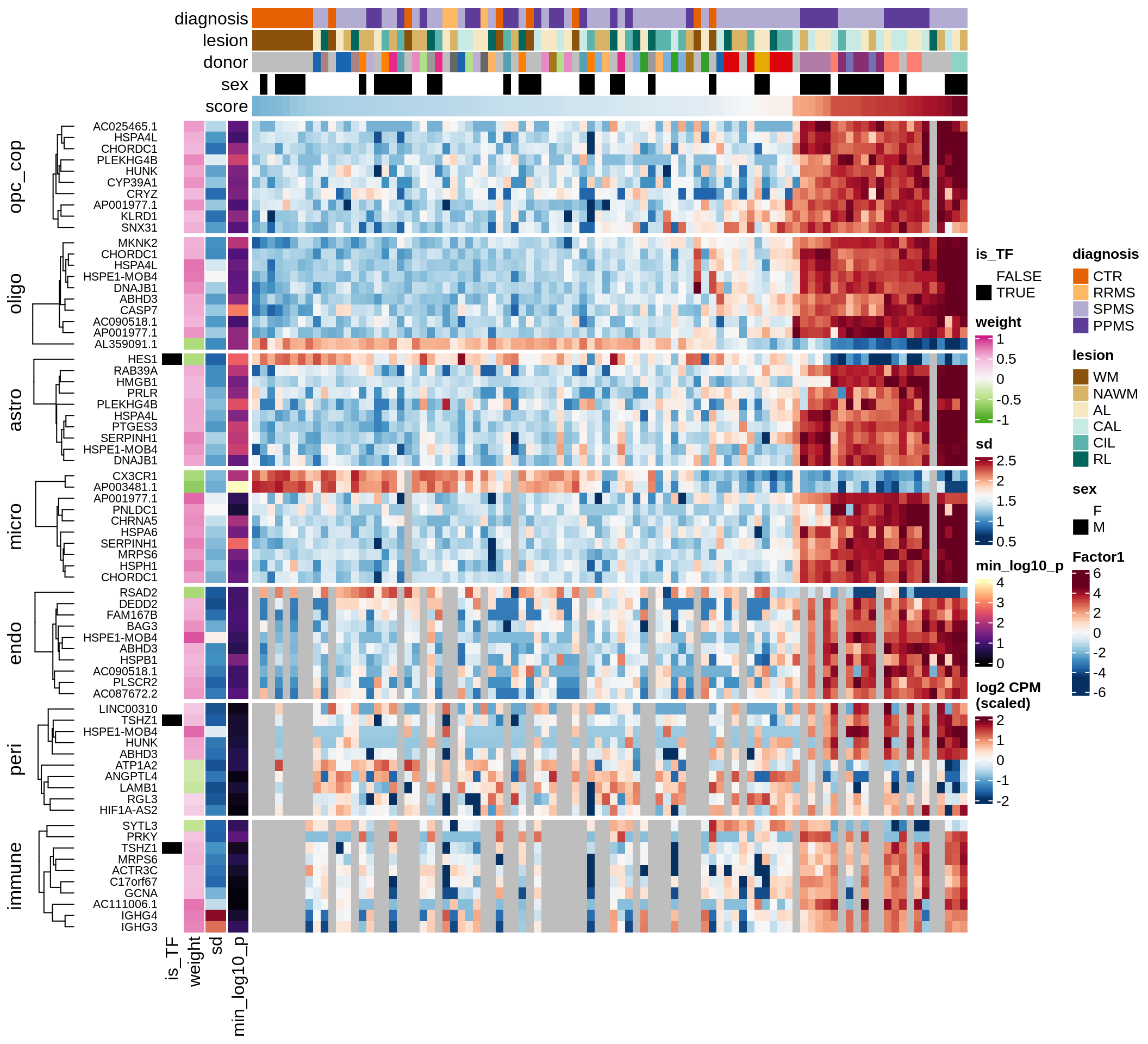
| Version | Author | Date |
|---|---|---|
| 7fb1b95 | wmacnair | 2021-11-25 |
G
Factor 2 top genes
include_graphics("figure/ms15_mofa_sample_wm_final_meta.Rmd/fig_factor2-1.png", error = FALSE)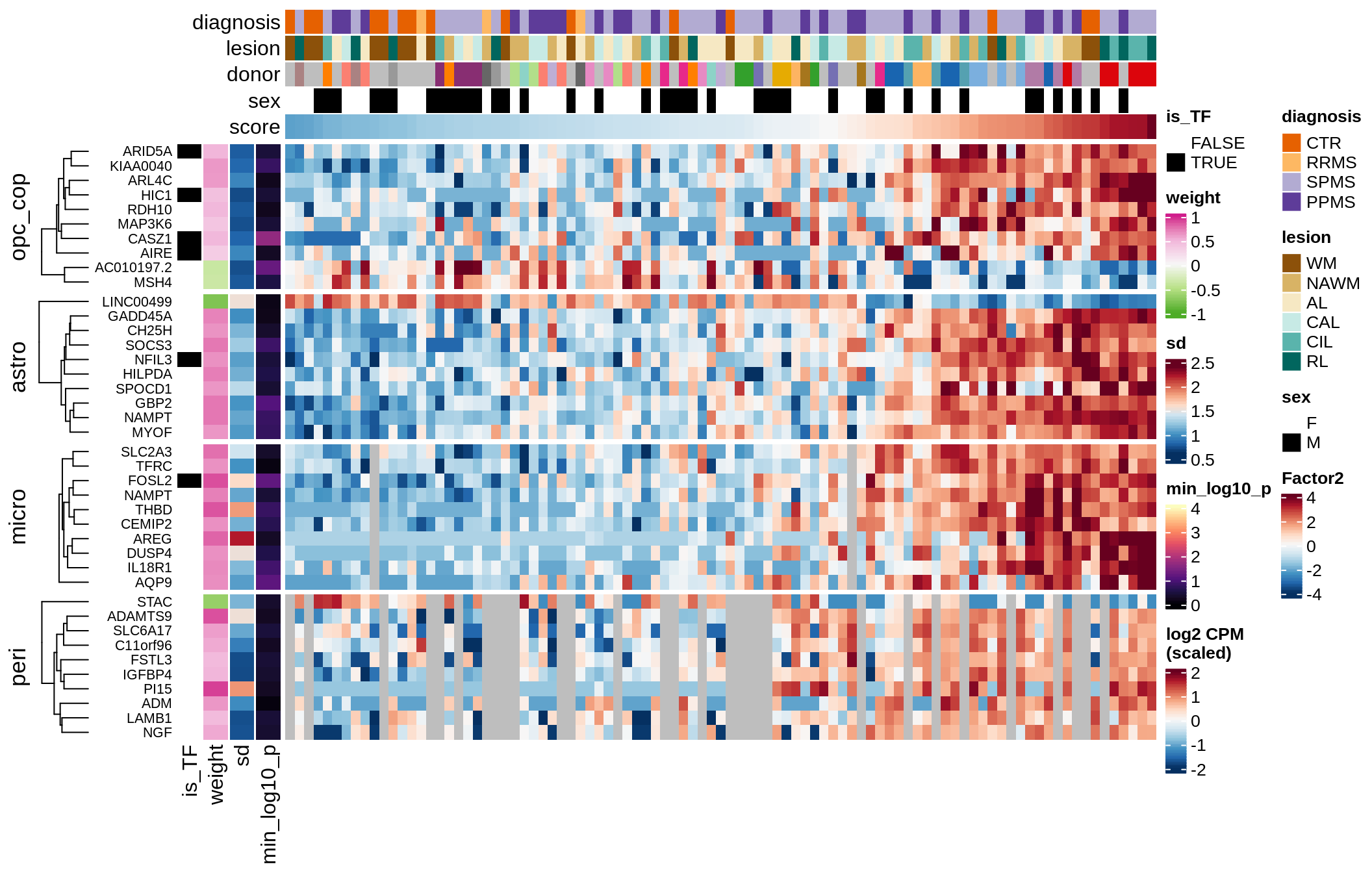
| Version | Author | Date |
|---|---|---|
| 7fb1b95 | wmacnair | 2021-11-25 |
Supplementary figures
S1
Contribution to variability in celltype abundances explained by lesion + patient in GM, no layers
include_graphics("figure/ms09_ancombc_mixed.Rmd/plot_lrt_results-2.png", error = FALSE)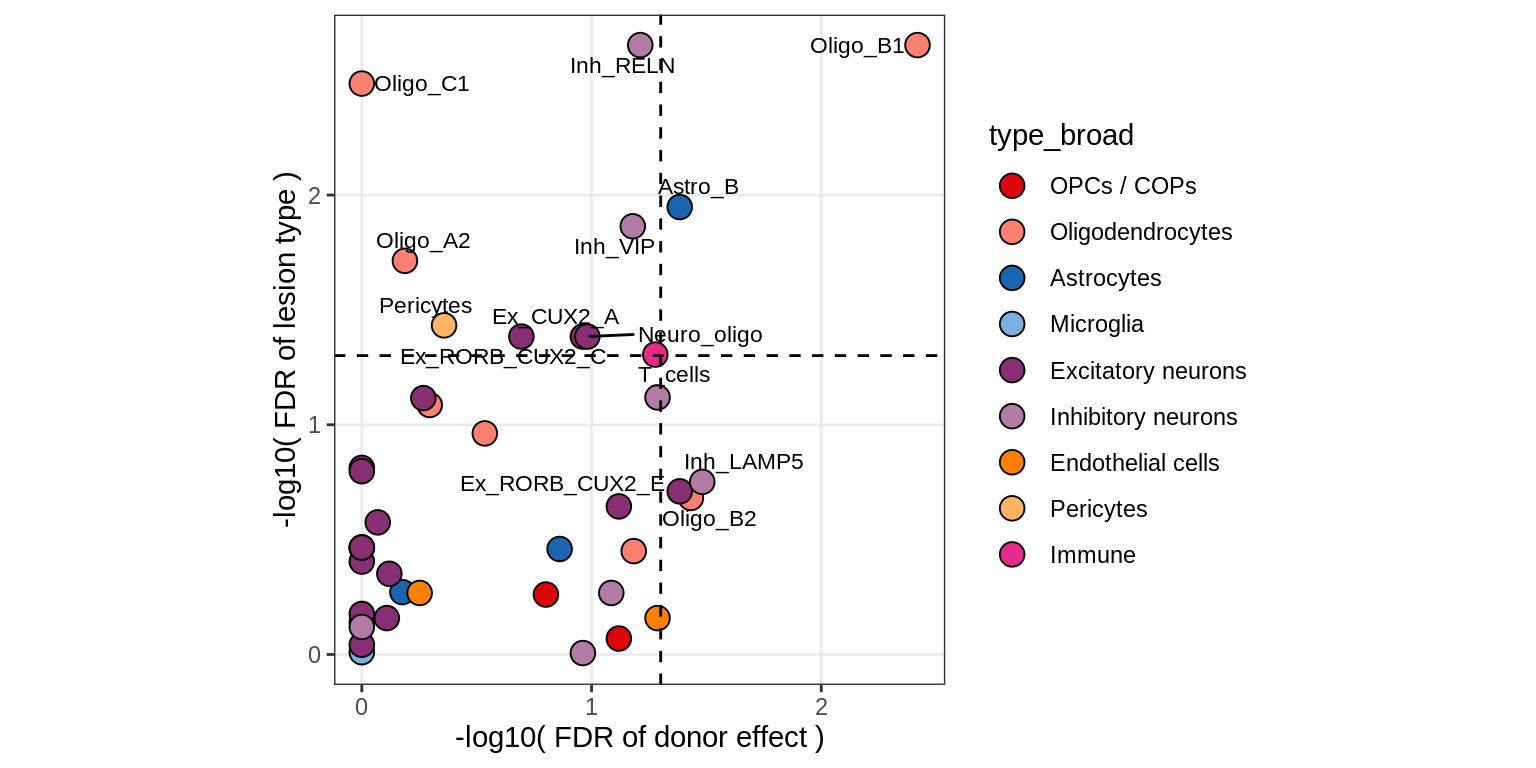
S2
Contribution to variability in celltype abundances explained by lesion + patient in GM, including 4 layer PCs
include_graphics("figure/ms09_ancombc_mixed.Rmd/plot_lrt_results-6.png", error = FALSE)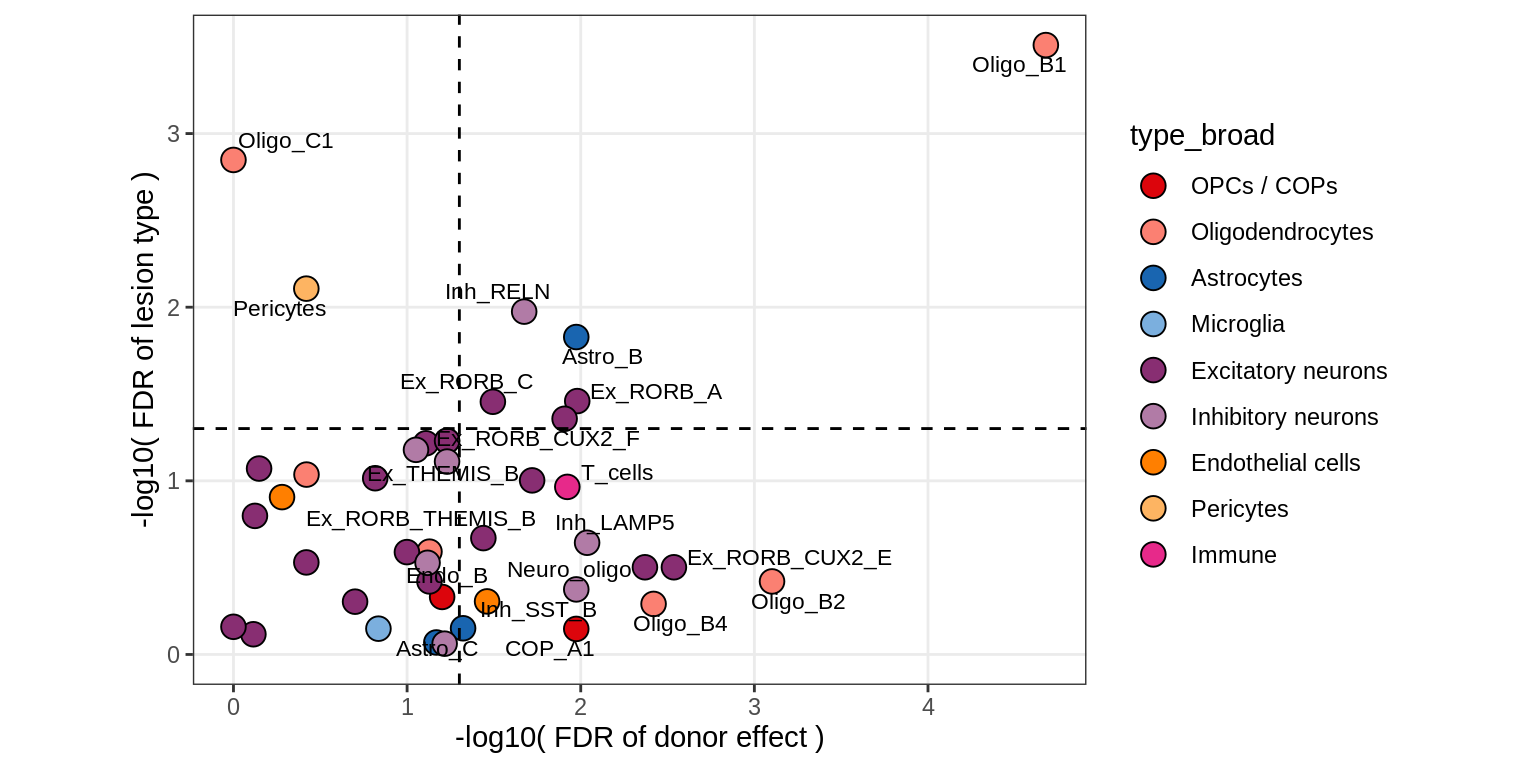
S3
DA results for GM, no layers
include_graphics("figure/ms09_ancombc_mixed.Rmd/plot_bootstraps_lesions-2.png", error = FALSE)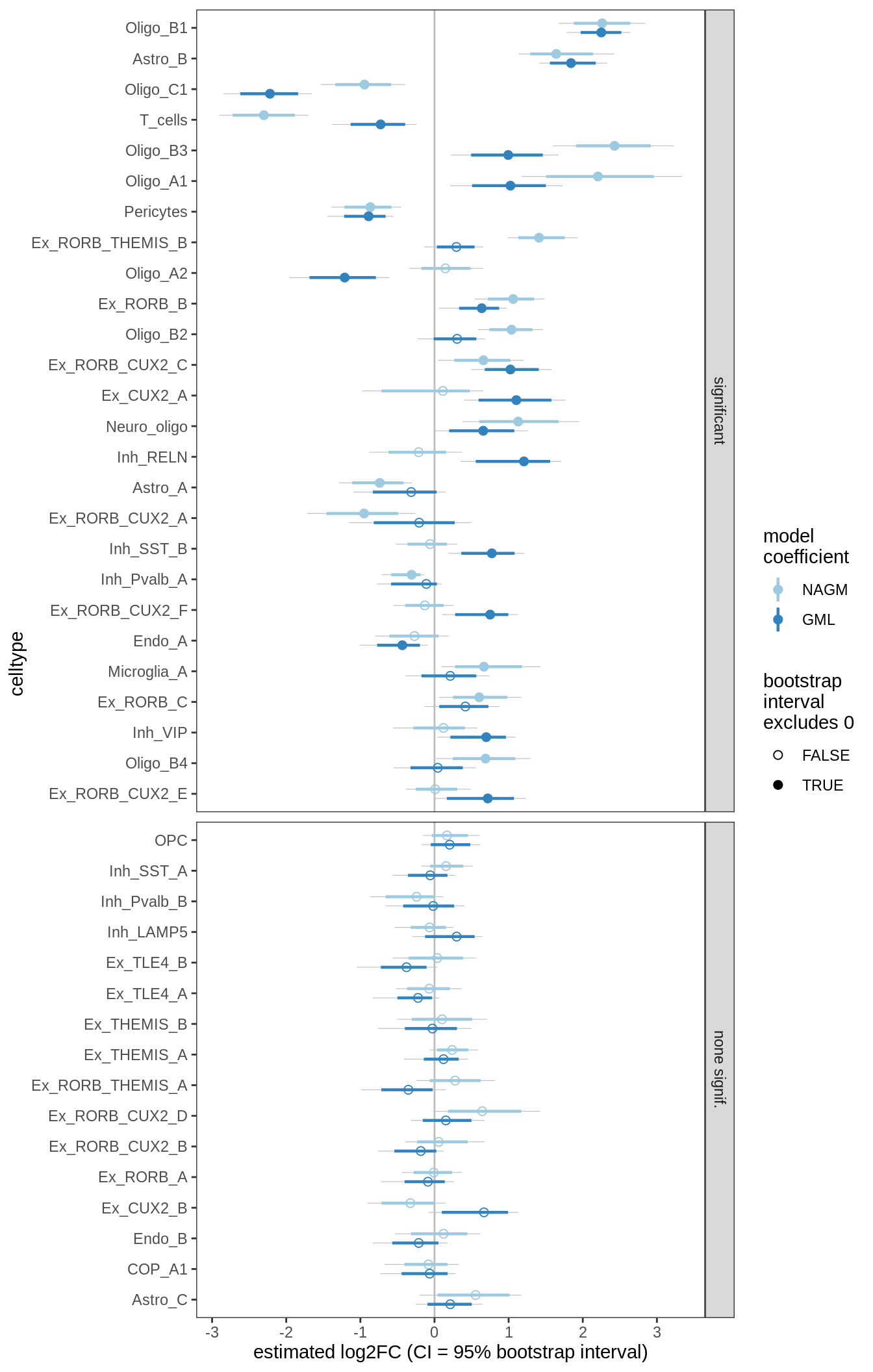
S4
DA results for GM, 4 layer PCs
include_graphics("figure/ms09_ancombc_mixed.Rmd/plot_bootstraps_lesions-6.png", error = FALSE)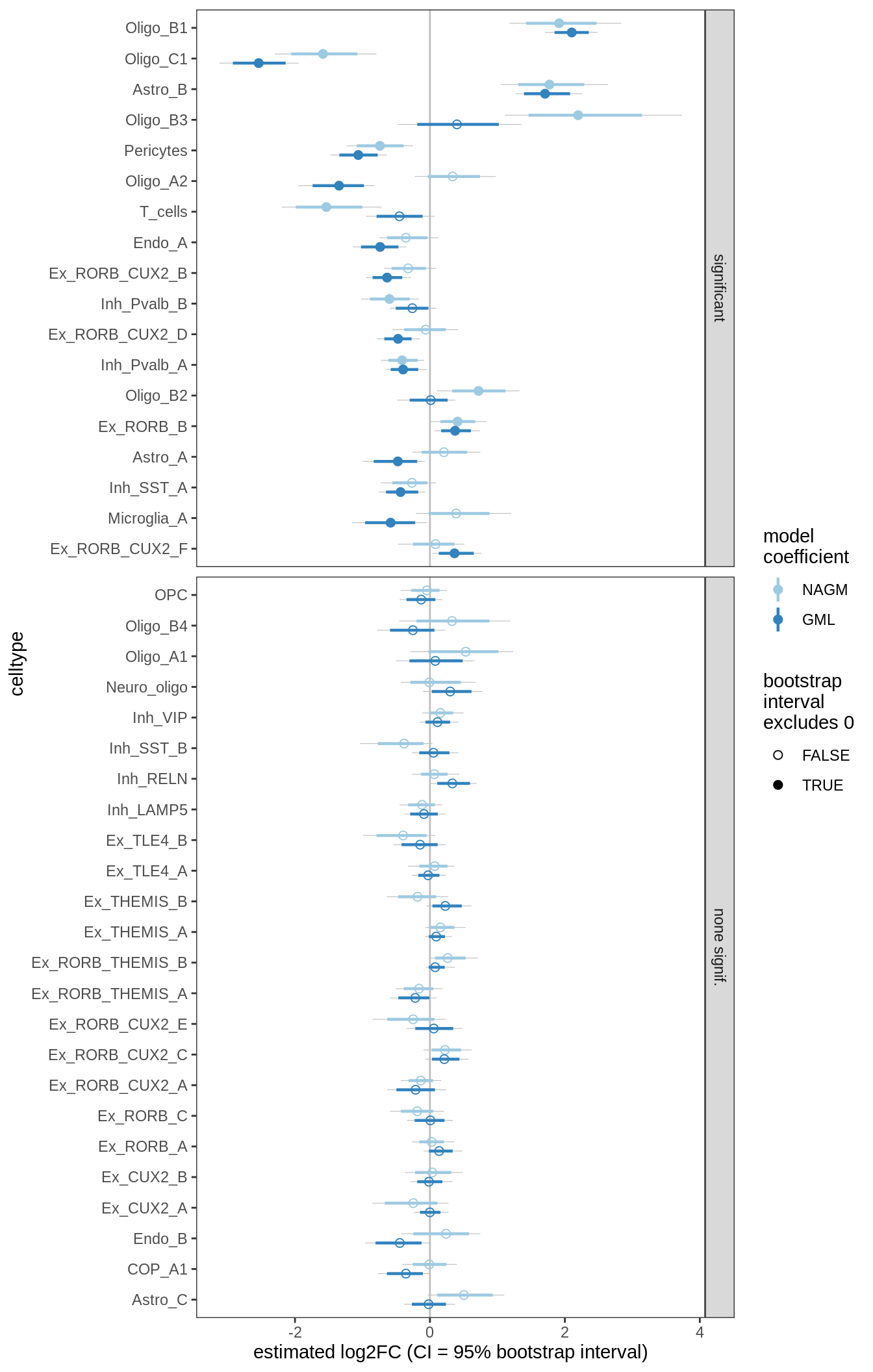
S5
Effect of including PCs, neurons
include_graphics("figure/ms09_ancombc_mixed.Rmd/plot_effect_of_pcs_lesions-1.png", error = FALSE)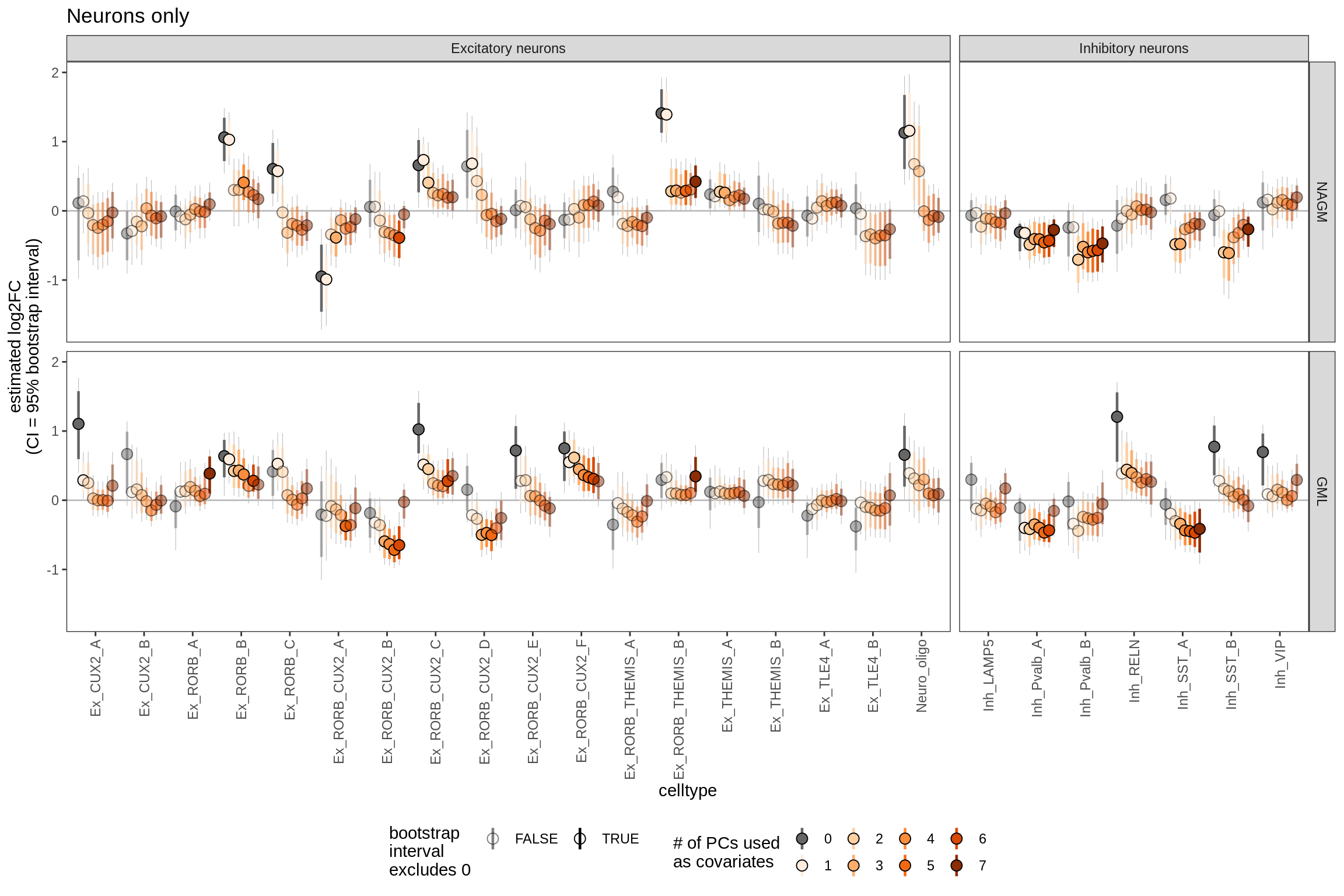
S6
Effect of including PCs, other celltypes
include_graphics("figure/ms09_ancombc_mixed.Rmd/plot_effect_of_pcs_lesions-2.png", error = FALSE)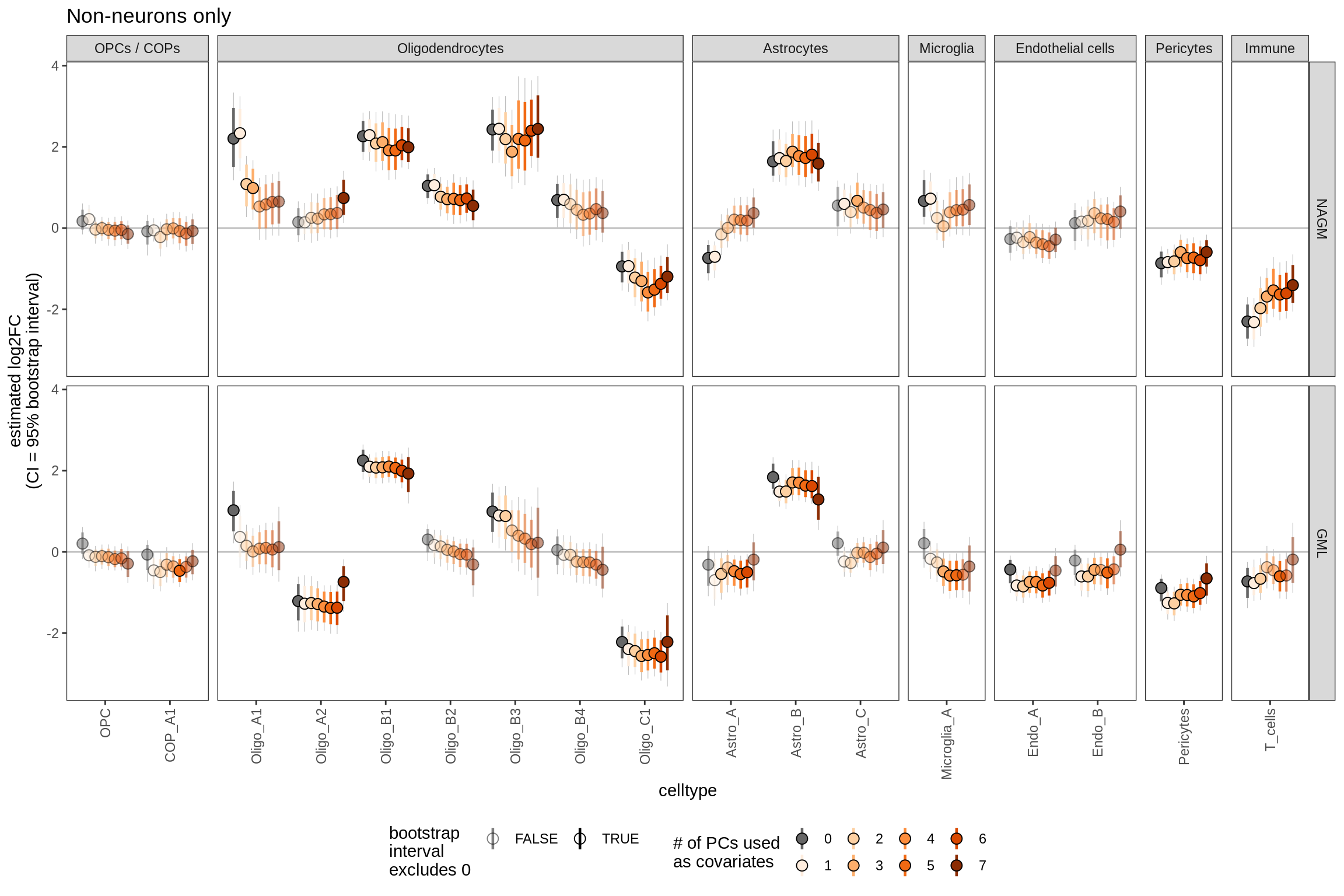
End
devtools::session_info()- Session info ---------------------------------------------------------------
setting value
version R version 4.0.5 (2021-03-31)
os CentOS Linux 7 (Core)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_US.UTF-8
ctype C
tz Europe/Zurich
date 2021-12-17
- Packages -------------------------------------------------------------------
package * version date lib source
assertthat * 0.2.1 2019-03-21 [2] CRAN (R 4.0.0)
BiocManager 1.30.16 2021-06-15 [1] CRAN (R 4.0.3)
BiocStyle * 2.18.1 2020-11-24 [1] Bioconductor
bslib 0.3.1 2021-10-06 [2] CRAN (R 4.0.5)
cachem 1.0.6 2021-08-19 [1] CRAN (R 4.0.5)
callr 3.7.0 2021-04-20 [2] CRAN (R 4.0.3)
cellranger 1.1.0 2016-07-27 [2] CRAN (R 4.0.0)
circlize * 0.4.13 2021-06-09 [1] CRAN (R 4.0.3)
cli 3.0.1 2021-07-17 [1] CRAN (R 4.0.3)
codetools 0.2-18 2020-11-04 [2] CRAN (R 4.0.3)
colorout * 1.2-2 2021-04-15 [1] Github (jalvesaq/colorout@79931fd)
colorspace 2.0-2 2021-06-24 [1] CRAN (R 4.0.3)
crayon 1.4.1 2021-02-08 [2] CRAN (R 4.0.3)
data.table * 1.14.2 2021-09-27 [2] CRAN (R 4.0.5)
DBI 1.1.1 2021-01-15 [2] CRAN (R 4.0.3)
desc 1.4.0 2021-09-28 [1] CRAN (R 4.0.5)
devtools 2.4.2 2021-06-07 [1] CRAN (R 4.0.3)
digest 0.6.28 2021-09-23 [2] CRAN (R 4.0.5)
dplyr 1.0.7 2021-06-18 [2] CRAN (R 4.0.3)
ellipsis 0.3.2 2021-04-29 [2] CRAN (R 4.0.3)
evaluate 0.14 2019-05-28 [2] CRAN (R 4.0.0)
fansi 0.5.0 2021-05-25 [2] CRAN (R 4.0.3)
fastmap 1.1.0 2021-01-25 [2] CRAN (R 4.0.3)
forcats * 0.5.1 2021-01-27 [2] CRAN (R 4.0.3)
fs 1.5.0 2020-07-31 [2] CRAN (R 4.0.2)
generics 0.1.1 2021-10-25 [2] CRAN (R 4.0.5)
ggplot2 * 3.3.5 2021-06-25 [1] CRAN (R 4.0.3)
git2r 0.28.0 2021-01-10 [1] CRAN (R 4.0.3)
GlobalOptions 0.1.2 2020-06-10 [1] CRAN (R 4.0.3)
glue 1.4.2 2020-08-27 [2] CRAN (R 4.0.3)
gridExtra 2.3 2017-09-09 [2] CRAN (R 4.0.0)
gtable 0.3.0 2019-03-25 [2] CRAN (R 4.0.0)
highr 0.9 2021-04-16 [2] CRAN (R 4.0.3)
htmltools 0.5.2 2021-08-25 [2] CRAN (R 4.0.5)
httpuv 1.6.3 2021-09-09 [2] CRAN (R 4.0.5)
jquerylib 0.1.4 2021-04-26 [2] CRAN (R 4.0.3)
jsonlite 1.7.2 2020-12-09 [2] CRAN (R 4.0.3)
knitr * 1.36 2021-09-29 [1] CRAN (R 4.0.5)
later 1.3.0 2021-08-18 [2] CRAN (R 4.0.5)
lifecycle 1.0.1 2021-09-24 [2] CRAN (R 4.0.5)
magrittr * 2.0.1 2020-11-17 [1] CRAN (R 4.0.3)
memoise 2.0.0 2021-01-26 [1] CRAN (R 4.0.3)
munsell 0.5.0 2018-06-12 [2] CRAN (R 4.0.0)
pillar 1.6.4 2021-10-18 [1] CRAN (R 4.0.5)
pkgbuild 1.2.0 2020-12-15 [1] CRAN (R 4.0.3)
pkgconfig 2.0.3 2019-09-22 [2] CRAN (R 4.0.0)
pkgload 1.2.3 2021-10-13 [2] CRAN (R 4.0.5)
prettyunits 1.1.1 2020-01-24 [2] CRAN (R 4.0.0)
processx 3.5.2 2021-04-30 [2] CRAN (R 4.0.3)
promises 1.2.0.1 2021-02-11 [2] CRAN (R 4.0.3)
ps 1.6.0 2021-02-28 [2] CRAN (R 4.0.3)
purrr 0.3.4 2020-04-17 [2] CRAN (R 4.0.0)
R6 2.5.1 2021-08-19 [2] CRAN (R 4.0.5)
RColorBrewer * 1.1-2 2014-12-07 [2] CRAN (R 4.0.0)
Rcpp 1.0.7 2021-07-07 [1] CRAN (R 4.0.3)
readxl * 1.3.1 2019-03-13 [2] CRAN (R 4.0.0)
remotes 2.4.1 2021-09-29 [1] CRAN (R 4.0.5)
rlang 0.4.12 2021-10-18 [2] CRAN (R 4.0.5)
rmarkdown 2.11 2021-09-14 [1] CRAN (R 4.0.5)
rprojroot 2.0.2 2020-11-15 [2] CRAN (R 4.0.3)
sass 0.4.0 2021-05-12 [2] CRAN (R 4.0.3)
scales * 1.1.1 2020-05-11 [2] CRAN (R 4.0.0)
sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.0.3)
shape 1.4.6 2021-05-19 [1] CRAN (R 4.0.1)
stringi 1.7.4 2021-08-25 [1] CRAN (R 4.0.5)
stringr * 1.4.0 2019-02-10 [2] CRAN (R 4.0.0)
testthat 3.1.0 2021-10-04 [2] CRAN (R 4.0.5)
tibble 3.1.5 2021-09-30 [1] CRAN (R 4.0.5)
tidyselect 1.1.1 2021-04-30 [2] CRAN (R 4.0.3)
usethis 2.1.2 2021-10-25 [1] CRAN (R 4.0.5)
utf8 1.2.2 2021-07-24 [1] CRAN (R 4.0.3)
vctrs 0.3.8 2021-04-29 [2] CRAN (R 4.0.3)
viridis * 0.6.2 2021-10-13 [1] CRAN (R 4.0.5)
viridisLite * 0.4.0 2021-04-13 [1] CRAN (R 4.0.1)
withr 2.4.2 2021-04-18 [2] CRAN (R 4.0.3)
workflowr * 1.6.2 2020-04-30 [1] CRAN (R 4.0.3)
xfun 0.27 2021-10-18 [1] CRAN (R 4.0.5)
yaml 2.2.1 2020-02-01 [2] CRAN (R 4.0.3)
[1] /pstore/home/macnairw/lib/conda_r3.12
[2] /pstore/home/macnairw/.conda/envs/r_4.0.3/lib/R/library
sessionInfo()R version 4.0.5 (2021-03-31)
Platform: x86_64-conda-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /pstore/home/macnairw/.conda/envs/r_4.0.3/lib/libopenblasp-r0.3.12.so
locale:
[1] LC_CTYPE=C LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] knitr_1.36 readxl_1.3.1 forcats_0.5.1 ggplot2_3.3.5
[5] scales_1.1.1 viridis_0.6.2 viridisLite_0.4.0 assertthat_0.2.1
[9] stringr_1.4.0 data.table_1.14.2 magrittr_2.0.1 circlize_0.4.13
[13] RColorBrewer_1.1-2 BiocStyle_2.18.1 colorout_1.2-2 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.7 prettyunits_1.1.1 ps_1.6.0
[4] rprojroot_2.0.2 digest_0.6.28 utf8_1.2.2
[7] R6_2.5.1 cellranger_1.1.0 evaluate_0.14
[10] highr_0.9 pillar_1.6.4 GlobalOptions_0.1.2
[13] rlang_0.4.12 callr_3.7.0 jquerylib_0.1.4
[16] rmarkdown_2.11 desc_1.4.0 devtools_2.4.2
[19] munsell_0.5.0 compiler_4.0.5 httpuv_1.6.3
[22] xfun_0.27 pkgconfig_2.0.3 pkgbuild_1.2.0
[25] shape_1.4.6 htmltools_0.5.2 tidyselect_1.1.1
[28] tibble_3.1.5 gridExtra_2.3 codetools_0.2-18
[31] fansi_0.5.0 crayon_1.4.1 dplyr_1.0.7
[34] withr_2.4.2 later_1.3.0 grid_4.0.5
[37] jsonlite_1.7.2 gtable_0.3.0 lifecycle_1.0.1
[40] DBI_1.1.1 git2r_0.28.0 cli_3.0.1
[43] stringi_1.7.4 cachem_1.0.6 remotes_2.4.1
[46] fs_1.5.0 promises_1.2.0.1 testthat_3.1.0
[49] bslib_0.3.1 ellipsis_0.3.2 generics_0.1.1
[52] vctrs_0.3.8 tools_4.0.5 glue_1.4.2
[55] purrr_0.3.4 pkgload_1.2.3 processx_3.5.2
[58] fastmap_1.1.0 yaml_2.2.1 colorspace_2.0-2
[61] BiocManager_1.30.16 sessioninfo_1.1.1 memoise_2.0.0
[64] usethis_2.1.2 sass_0.4.0