Reformatting UKB ‘all_lkps_maps.xlsx’
Alasdair Warwick
17 February, 2022
Last updated: 2022-02-17
Checks: 5 2
Knit directory: codemapper/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210923) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| ~/Documents/PhDLocal/codemapper/ukbb_pan_ancestry-master/data/UKB_PHENOME_ICD9_PHECODE_MAP_20200109.txt | ukbb_pan_ancestry-master/data/UKB_PHENOME_ICD9_PHECODE_MAP_20200109.txt |
| ~/Documents/PhDLocal/codemapper/ukbb_pan_ancestry-master/data/UKB_PHENOME_ICD10_PHECODE_MAP_20200109.txt | ukbb_pan_ancestry-master/data/UKB_PHENOME_ICD10_PHECODE_MAP_20200109.txt |
<<<<<<< HEAD Repository version: eb7af13
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
<<<<<<< HEAD The results in this page were generated with repository version eb7af13. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files. ======= The results in this page were generated with repository version a44eb90. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files. >>>>>>> dev_cy
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Renviron
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: _targets/
Ignored: all_lkps_maps.db
Ignored: renv/library/
Ignored: renv/staging/
Ignored: tar_make.R
<<<<<<< HEAD
Unstaged changes:
Modified: .Rbuildignore
Modified: R/constants.R
Modified: R/utils.R
Modified: _targets.R
Modified: man/build_all_lkps_maps.Rd
=======
Untracked files:
Untracked: ukbb_pan_ancestry-master/
Unstaged changes:
Modified: analysis/phecodes.Rmd
>>>>>>> dev_cy
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/phecodes.Rmd) and HTML (public/phecodes.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 81047b4 | rmgpanw | 2022-02-17 | setup for gitlab CI with pkgdown site and test coverage; start adding read3 to snomed mapping |
| Rmd | 5c2a3e3 | Chuin Ying Ung | 2022-02-17 | update _targets.R (housekeeping) and phecode.Rmd |
| html | 5c2a3e3 | Chuin Ying Ung | 2022-02-17 | update _targets.R (housekeeping) and phecode.Rmd |
| Rmd | 5c2a3e3 | Chuin Ying Ung | 2022-02-17 | update _targets.R (housekeeping) and phecode.Rmd |
| html | 5c2a3e3 | Chuin Ying Ung | 2022-02-17 | update _targets.R (housekeeping) and phecode.Rmd |
| Rmd | 8d14804 | Chuin Ying Ung | 2022-02-17 | add phecode.Rmd |
Overview
Aim: to explore the phecode lookup and mapping files.
Background - phecodes
- LINKS TO KEY PUBLICATIONS AND WEBPAGES
- WHAT CODING SYSTEM IS PHECODES BASED ON, AND WHICH CLINICAL CODES CAN BE MAPPED TO THIS?
PheWAS R package
- READ HELP PAGE FROM RUNNING BELOW
- TO START, PERHAPS TRY RUNNING THE EXAMPLE (AT BOTTOM OF HELP PAGE) - COPY AND PASTE INTO THIS NOTEBOOK SO WE CAN SEE THE OUTPUT AT EACH STEP
Generate some example data
#Create the phecode table- translates the codes, adds exclusions, and reshapes to a wide format. #Sum up the counts in the data where applicable.
#Combine the data
#Run the PheWAS
#Plot the results 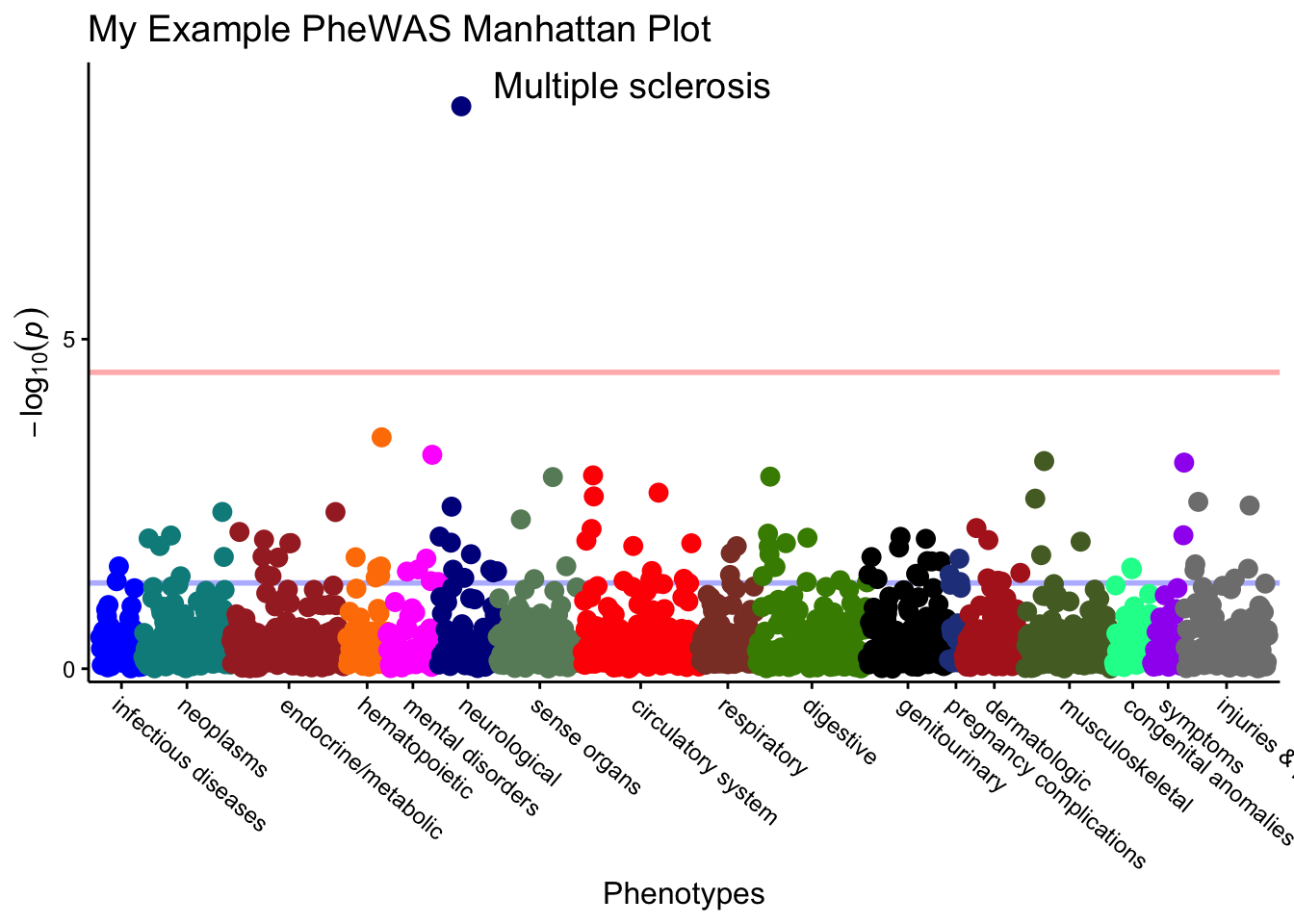
#Add PheWAS descriptions
#List the top 10 results
phenotype snp covariates beta SE OR
549 335 rsEXAMPLE sex 0.3967548 0.06684421 1.4869913
431 288.2 rsEXAMPLE sex 1.3847304 0.38379930 3.9937489
507 315.1 rsEXAMPLE sex 1.4280228 0.41430554 4.1704451
1475 716.8 rsEXAMPLE sex 1.3794156 0.40731579 3.9725795
1686 798.1 rsEXAMPLE sex 1.0432921 0.30938935 2.8385463
743 401.2 rsEXAMPLE sex 0.8005488 0.24653912 2.2267626
1002 525.1 rsEXAMPLE sex 0.9682267 0.29931207 2.6332707
680 375.1 rsEXAMPLE sex -1.2438354 0.38503286 0.2882764
838 440.2 rsEXAMPLE sex 0.6394982 0.20822694 1.8955294
744 401.21 rsEXAMPLE sex 1.2694273 0.41871628 3.5588138
p type n_total n_cases n_controls HWE_p allele_freq
549 2.929243e-09 logistic 4404 1873 2531 1 0.5128292
431 3.086207e-04 logistic 4731 26 4705 1 0.5110970
507 5.672965e-04 logistic 4762 22 4740 1 0.5115498
1475 7.076450e-04 logistic 4243 23 4220 1 0.5103700
1686 7.459717e-04 logistic 4973 42 4931 1 0.5115624
743 1.165681e-03 logistic 4924 70 4854 1 0.5111698
1002 1.217109e-03 logistic 4284 46 4238 1 0.5109711
680 1.235888e-03 logistic 4501 27 4474 1 0.5118862
838 2.132291e-03 logistic 4108 102 4006 1 0.5139971
744 2.431709e-03 logistic 4876 22 4854 1 0.5105619
n_no_snp formula expanded_formula
549 0 `335` ~ `rsEXAMPLE` + `sex` (Intercept) + rsEXAMPLE + sexM
431 0 `288.2` ~ `rsEXAMPLE` + `sex` (Intercept) + rsEXAMPLE + sexM
507 0 `315.1` ~ `rsEXAMPLE` + `sex` (Intercept) + rsEXAMPLE + sexM
1475 0 `716.8` ~ `rsEXAMPLE` + `sex` (Intercept) + rsEXAMPLE + sexM
1686 0 `798.1` ~ `rsEXAMPLE` + `sex` (Intercept) + rsEXAMPLE + sexM
743 0 `401.2` ~ `rsEXAMPLE` + `sex` (Intercept) + rsEXAMPLE + sexM
1002 0 `525.1` ~ `rsEXAMPLE` + `sex` (Intercept) + rsEXAMPLE + sexM
680 0 `375.1` ~ `rsEXAMPLE` + `sex` (Intercept) + rsEXAMPLE + sexM
838 0 `440.2` ~ `rsEXAMPLE` + `sex` (Intercept) + rsEXAMPLE + sexM
744 0 `401.21` ~ `rsEXAMPLE` + `sex` (Intercept) + rsEXAMPLE + sexM
note description group
549 Multiple sclerosis neurological
431 Elevated white blood cell count hematopoietic
507 Learning disorder mental disorders
1475 Palindromic rheumatism musculoskeletal
1686 Chronic fatigue syndrome symptoms
743 Hypertensive heart and/or renal disease circulatory system
1002 Loss of teeth or edentulism digestive
680 Dry eyes sense organs
838 Atherosclerosis of the extremities circulatory system
744 Hypertensive heart disease circulatory system#Create a nice interactive table (eg, in RStudio)
Mapping files
DOWNLOAD PHECODE MAPPING FILES TO DATA DIRECTORY,THEN LOAD INTO R
<<<<<<< HEAD
How many ICD-10 codes are included in the UK Biobank lookup table? (head shown below)
# A tibble: 17,934 × 12
ICD10_CODE ALT_CODE USAGE USAGE_UK DESCRIPTION MODIFIER_4 MODIFIER_5
<chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 A00 A00 DEFAULT 3 Cholera <NA> <NA>
2 A00.0 A000 DEFAULT 3 Cholera due to Vi… <NA> <NA>
3 A00.1 A001 DEFAULT 3 Cholera due to Vi… <NA> <NA>
4 A00.9 A009 DEFAULT 3 Cholera, unspecif… <NA> <NA>
5 A01 A01 DEFAULT 3 Typhoid and parat… <NA> <NA>
6 A01.0 A010 DEFAULT 3 Typhoid fever <NA> <NA>
7 A01.1 A011 DEFAULT 3 Paratyphoid fever… <NA> <NA>
8 A01.2 A012 DEFAULT 3 Paratyphoid fever… <NA> <NA>
9 A01.3 A013 DEFAULT 3 Paratyphoid fever… <NA> <NA>
10 A01.4 A014 DEFAULT 3 Paratyphoid fever… <NA> <NA>
# … with 17,924 more rows, and 5 more variables: QUALIFIERS <chr>,
# GENDER_MASK <chr>, MIN_AGE <chr>, MAX_AGE <chr>, TREE_DESCRIPTION <chr>- Which ICD-10 codes map to >1 phecode? (I think this is mentioned on the phecode website, and in Spiros’ github repo readme)
Downloaded files from ukbb pan ancestry https://github.com/atgu/ukbb_pan_ancestry
How many ICD codes are included in the UK Biobank lookup table? (head shown below)
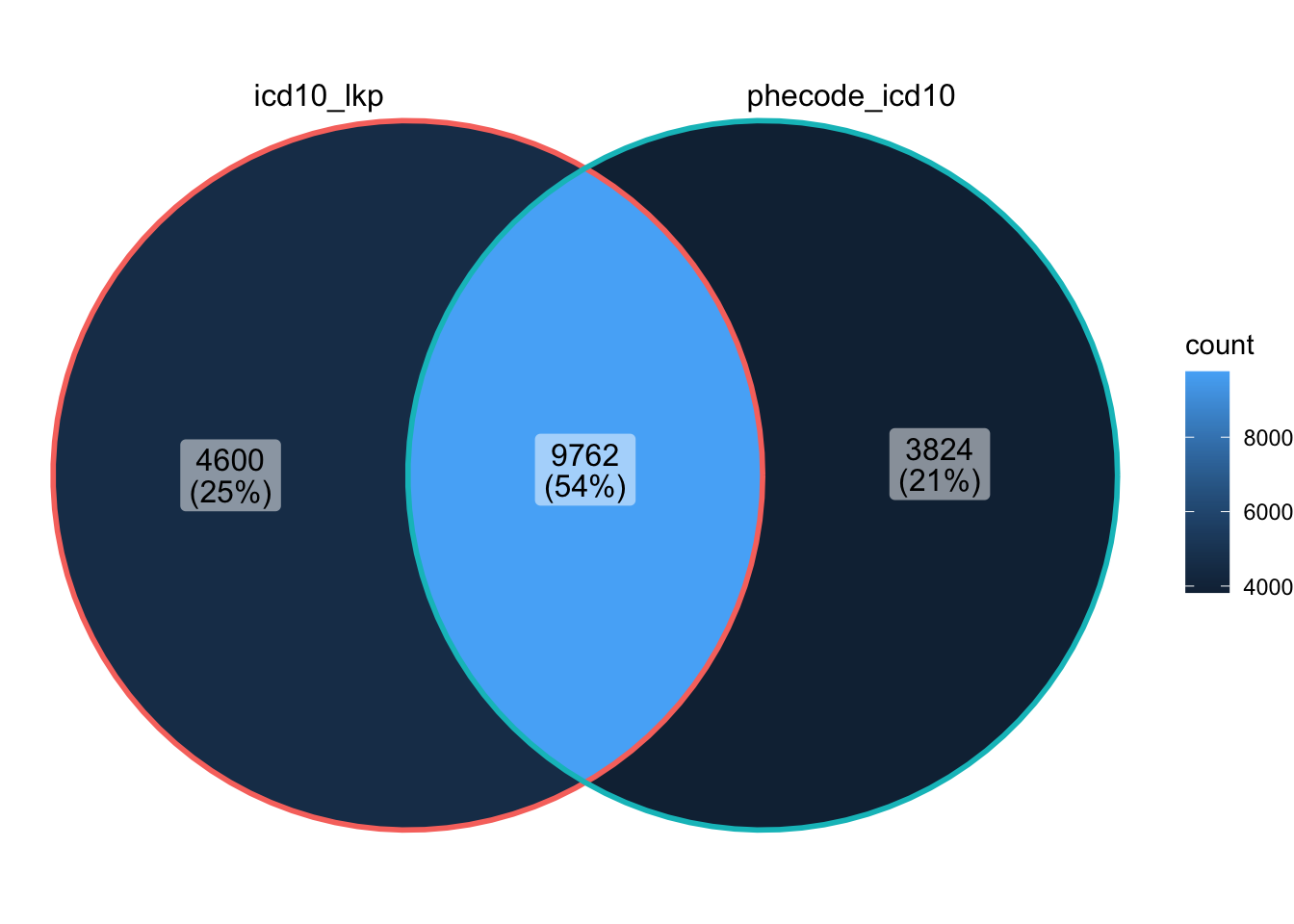
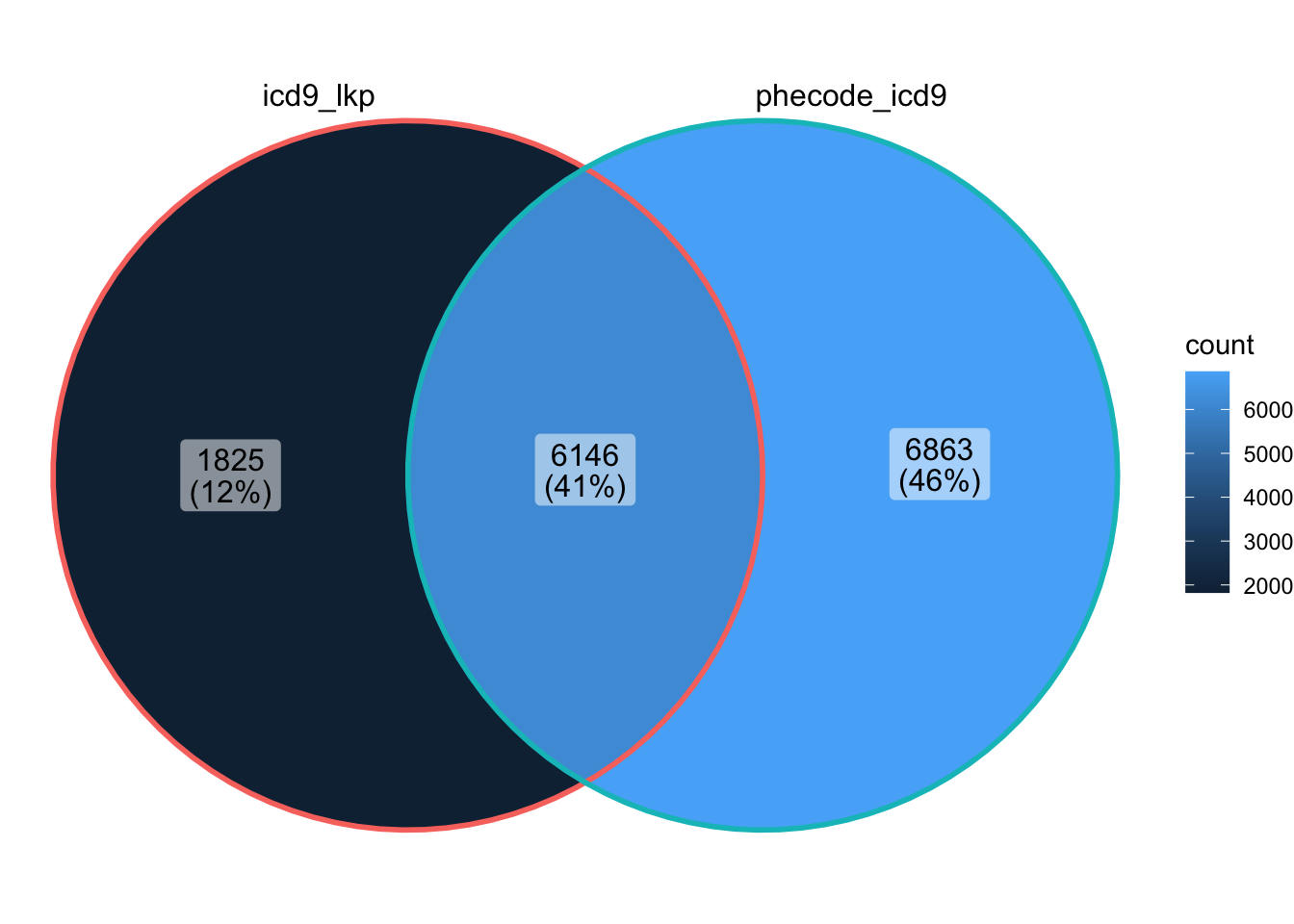
Which ICD-10 codes are not shared by phecode_map and all_lkps_maps
How many ICD-10 codes map to >1 phecode? (I think this is mentioned on the phecode website, and in Spiros’ github repo readme)
[1] 7017[1] 1004- How many ICD-9 codes map to >1 phecode?
[1] 6917[1] 2772
R version 4.1.0 (2021-05-18)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRblas.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRlapack.dylib
locale:
[1] en_GB.UTF-8/en_GB.UTF-8/en_GB.UTF-8/C/en_GB.UTF-8/en_GB.UTF-8
attached base packages:
<<<<<<< HEAD
[1] parallel stats graphics grDevices datasets utils methods
[8] base
other attached packages:
[1] PheWAS_0.99.5-5 codemapper_0.0.0.9000 ukbwranglr_0.0.0.9000
[4] targets_0.8.0 crosstalk_1.1.1 readxl_1.3.1
[7] reactable_0.2.3 forcats_0.5.1 stringr_1.4.0
[10] dplyr_1.0.7 purrr_0.3.4 readr_2.0.2
[13] tidyr_1.1.4 tibble_3.1.4 ggplot2_3.3.5
[16] tidyverse_1.3.1 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] minqa_1.2.4 colorspace_2.0-2 ellipsis_0.3.2
[4] rprojroot_2.0.2 fs_1.5.0 rstudioapi_0.13
[7] mice_3.14.0 ggrepel_0.9.1 DT_0.20
[10] fansi_0.5.0 lubridate_1.7.10 mathjaxr_1.4-0
[13] xml2_1.3.2 codetools_0.2-18 splines_4.1.0
[16] knitr_1.34 jsonlite_1.7.2 nloptr_2.0.0
[19] logistf_1.24.1 broom_0.7.9 dbplyr_2.1.1
[22] shiny_1.7.0 compiler_4.1.0 httr_1.4.2
[25] backports_1.2.1 assertthat_0.2.1 Matrix_1.3-3
[28] fastmap_1.1.0 cli_3.0.1 later_1.3.0
[31] htmltools_0.5.2 tools_4.1.0 igraph_1.2.6
[34] gtable_0.3.0 glue_1.4.2 Rcpp_1.0.7
[37] cellranger_1.1.0 jquerylib_0.1.4 vctrs_0.3.8
[40] nlme_3.1-152 lmtest_0.9-39 xfun_0.24
[43] ps_1.6.0 lme4_1.1-28 rvest_1.0.1
[46] mime_0.12 CompQuadForm_1.4.3 lifecycle_1.0.1
[49] renv_0.13.2 formula.tools_1.7.1 MASS_7.3-54
[52] zoo_1.8-9 scales_1.1.1 hms_1.1.1
[55] promises_1.2.0.1 meta_5.2-0 metafor_3.0-2
[58] yaml_2.2.1 sass_0.4.0 stringi_1.7.4
[61] highr_0.9 boot_1.3-28 operator.tools_1.6.3
[64] rlang_0.4.11 pkgconfig_2.0.3 evaluate_0.14
[67] lattice_0.20-44 htmlwidgets_1.5.4 processx_3.5.2
[70] tidyselect_1.1.1 magrittr_2.0.1 R6_2.5.1
[73] generics_0.1.0 DBI_1.1.1 pillar_1.6.3
[76] haven_2.4.3 whisker_0.4 withr_2.4.3
[79] mgcv_1.8-35 survival_3.2-13 modelr_0.1.8
[82] crayon_1.4.1 utf8_1.2.2 tzdb_0.1.2
[85] rmarkdown_2.11 grid_4.1.0 data.table_1.14.2
[88] callr_3.7.0 git2r_0.28.0 reprex_2.0.1
[91] digest_0.6.28 xtable_1.8-4 httpuv_1.6.3
[94] munsell_0.5.0 bslib_0.3.1