ROI-generation
ROI-generation
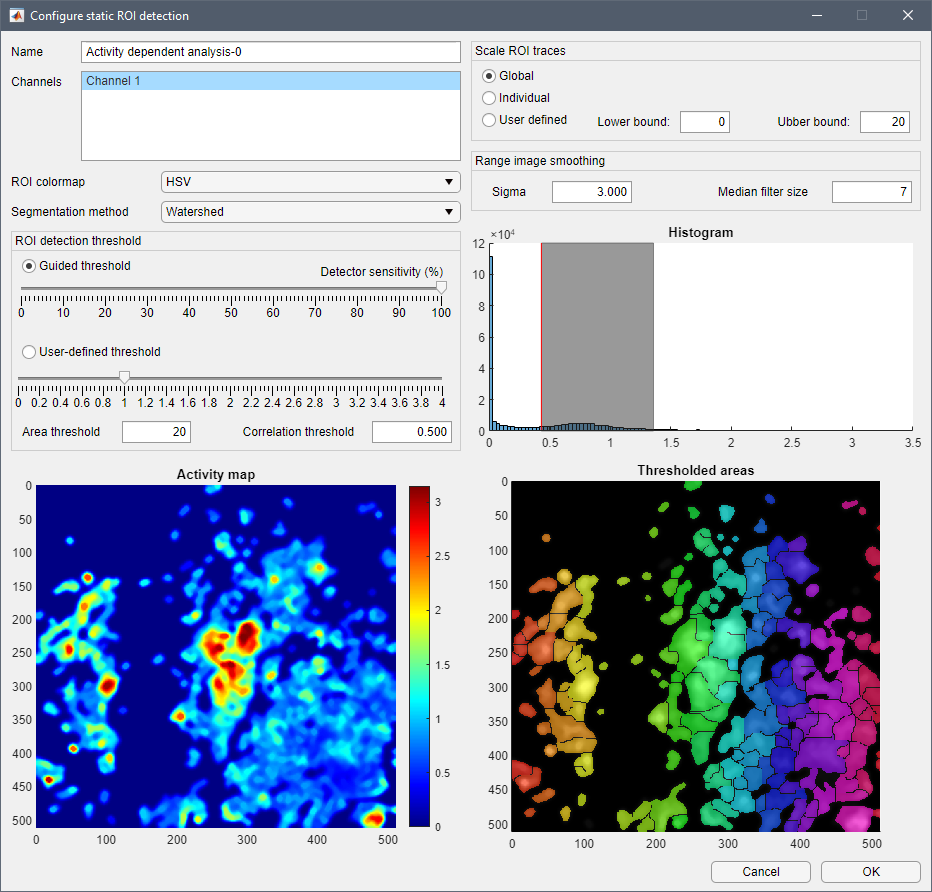
MSparkles offers several ways to create ROIs, called Analysis modes. A core feature of MSparkles is automatic ROI detection. In addition to that, MSparkles offers ROI-grid and Global ROI analyses. Of course, it is also possible to manually dra ROIs, or import a set of ImageJ ROIs. Analysis modes come with the ability to be intersected with each other to create differential analyses of, e.g. cell somata and gliapil/neuropil.
 ROI grid
ROI grid Global ROI
Global ROI Manual ROIs
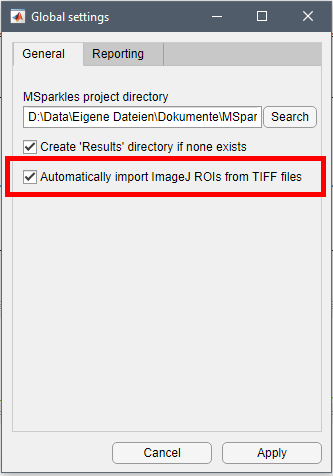
Manual ROIs Import ImageJ ROIs
Import ImageJ ROIs